Abstract
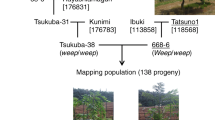
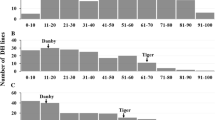
Most Japanese chestnuts have difficult-peeling pellicles, which make it time-consuming and costly to remove the pellicle from the nut during processing. Therefore, an easy-peeling pellicle has been an important goal of Japanese chestnut breeding programs. In previous studies, we identified the peeling locus, which controls peelability in Japanese chestnut. Homozygous recessive plants (p/p) have an easy-peeling pellicle. Here, we developed a novel simple sequence repeat (SSR) marker, CmSca06716, which is suitable for the selection of pellicle peelability in Japanese chestnut. We also constructed a fine map of Tsukuba-43 and localized the peeling locus within a 1.1-cM region between SSR markers CmSca05637 (or CmSca01731) and CmSca03291. The CmSca06716 marker co-segregated with the peeling locus in all F1 plantlets of a ‘Porotan’ × Tsukuba-43 cross. The peeling region was located within a 1.5-Mb region of chromosome 1 in two Chinese chestnut genome assemblies. The 394-bp allele of CmSca06716 was linked in the coupling phase to the recessive easy-peeling allele (p) possessed by ’Porotan’, ’Porosuke’, and ‘Yakko’, which are easy to peel, and in their relatives. We also identified three new cultivars that were heterozygous for the 394-bp allele at the CmSca06716 locus and the p allele at the peeling locus. Furthermore, we developed a cost-efficient and rapid marker-assisted selection system and applied it to our breeding program for Japanese chestnut. This information will lead to a better understanding of the genetic control and physiological mechanisms underlying pellicle peelability in Japanese chestnut.



Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The name, version, and parameters of the software used in this study are described in the “Materials and methods” section.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bounous G, Marinoni DT (2004) Chestnut: botany, horticulture, and utilization. Hortic Rev 31:291–347
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1010
Brumlop S, Reichenbecher W, Tappeser B, Finckh MR (2013) What is the SMARTest way to breed plants and increase agrobiodiversity? Euphytica 194:53–66
Buck EJ, Hadonou M, James CJ, Blakesley D, Russell K (2003) Isolation and characterization of polymorphic microsatellites in European chestnut (Castanea sativa Mill.). Mol Ecol Notes 3:239–241
Darvasi A, Soller M (1992) Selective genotyping for determination of linkage between a marker locus and a quantitative trait locus. Theor Appl Genet 85:353–359
Grattapaglia D, Sederoff R (1994) Genetic-linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Inoue E, Ning L, Hara H, Ruan SA, Anzai H (2009) Development of simple sequence repeat markers in Chinese chestnut and their characterization in diverse chestnut cultivars. J Am Soc Hortic Sci 134:610–617
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugenic 12:172–175
Kotobuki K (1994) Chestnut. In: Matsuo T (ed.) Collected data of plant genetic resources., Kodansya, Yokyo, pp. 1174–1184 (In Japanese)
Luby JJ, Shaw DV (2001) Does marker-assisted selection make dollars and sense in a fruit breeding program? HortScience 36:872–879
Marinoni D, Akkak A, Bounous G, Edwards KJ, Botta R (2003) Development and characterization of microsatellite markers in Castanea sativa (Mill.). Mol Breeding 11:127–136
Nishio S, Yamamoto T, Terakami S, Sawamura Y, Takada N, Nishitani C, Saito T (2011) Novel genomic and EST-derived SSR markers in Japanese chestnuts. Sci Hort 130:838–846
Nishio S, Takada N, Yamamoto T, Terakami S, Hayashi T, Sawamura Y, Saito T (2013) Mapping and pedigree analysis of the gene that controls the easy peel pellicle trait in Japanese chestnut (Castanea crenata Sieb. et Zucc.). Tree Genet Genomes 9:723–730
Nishio S, Iketani H, Fujii H, Yamamoto T, Terakami S, Takada N, Saito T (2014) Use of population structure and parentage analyses to elucidate the spread of native cultivars of Japanese chestnut. Tree Genet Genomes 10:1171–1180
Nishio S, Terakami S, Matsumoto T, Yamamoto T, Takada N, Kato H, Katayose Y, Saito T (2018) Identification of QTLs for agronomic traits in the Japanese chestnut (Castanea crenata Sieb. et Zucc.) Breeding. Horticult J 87:43–54
Pereira-Lorenzo S, Ballester A, Corredoira E, Vieitez AM, Agnanostakis S, Costa R, Bounous G, Botta R, Beccaro GL, Kubisiak TL, Conedera M et al (2012) Chestnut. In: Badenes MJ, Byrne DH (eds) Fruit breeding. Springer, New York, pp 729–769
Saito T, Kotobuki K, Sawamura Y, Abe K, Terai O, Shoda M, Takada N, Sato Y, Hirabayashi T, Sato A, Nishibata T et al (2009) New Japanese chestnut cultivar ‘Porotan.’ Bull Natl Inst Fruit Tree Sci 9:1–9 (In Japanese with English abstract)
Saito T, Takada N, Sawamura Y, Nishio S, Hirabayashi T, Sato A, Kato H, Onoue N, Uchida M (2021) New Japanese chestnut cultivar ‘Porosuke.’ J NARO Res Dev 7:39–46 (In Japanese with English abstract)
Sato A, Sawamura Y, Takada N, Hirabayashi T (2008) Relationship between inbreeding coefficients and plant height of 1-year-old seedlings in crosses among Japanese pear (Pyrus pyrifolia Nakai) cultivars/selections. Sci Hort 117:85–88
Sato A, Tanaka K, Takada N, Sawamura Y, Hirabayashi T (2010) Comparison of phenolic content of easily removed pellicle of Japanese chestnut ‘Porotan’ with other Japanese and Chinese chestnut cultivars. J Japan Soc Hor Sci 79:258–262
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Shirasawa K, Nishio S, Terakami S, Botta R, Marinoni DT, Isobe S (2021) Chromosome-level genome assembly of Japanese chestnut (Castanea crenata Sieb. et Zucc.) reveals conserved chromosomal segments in woody rosids. DNA Research 28: dsab016.
Shoda M, Takada N, Saito T, Sawamura Y, Kotobuki K (2006) A method for quickly removing pellicles from chestnuts by deep frying cooking oil. Bull Natl Inst Fruit Tree Sci 5:21–27 (In Japanese with English abstract)
Staton M, Zhebentyayeva T, Olukolu B, Fang GC, Nelson D, Carlson JE, Abbott AG (2015) Substantial genome synteny preservation among woody angiosperm species: comparative genomics of Chinese chestnut (Castanea mollissima) and plant reference genomes. BMC Genomics 16:744
Sun YS, Lu ZQ, Zhu XF, Ma H (2020) Genomic basis of homoploid hybrid speciation within chestnut trees. Nat Commun 11:3375
Takada N, Nishio S, Yamada M, Sawamura Y, Sato A, Hirabayashi T, Saito T (2012) Inheritance of the easy-peeling pellicle trait of Japanese chestnut cultivar Porotan. HortScience 47:845–847
Takada N, Yamada M, Nishio S, Sawamura Y, Sato A, Onoue N, Saito T (2017) Existence of genetic differences in pellicle peelability in Japanese chestnut (Castanea crenata Sieb. et Zucc.) cultivars and selections with difficult-peeling pellicles. Horticult J 86:456–462
Takada N, Yamada M, Nishio S, Kato H, Sawamura Y, Sato A, Onoue N, Saito T (2018) The investigation of pellicle peelability on Japanese chestnut cultivar of ‘Yakko’ (Castanea crenata Sieb. et Zucc.). Sci Hort 234:146–151
Tan SC, Yiap BC (2009) DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol 2009:574398. https://doi.org/10.1155/2009/574398
Terakami S, Adachi Y, Takeuchi Y, Takada N, Nishio S, Saito T, Yamamoto T (2021) Development of an SSR marker set for efficient selection for resistance to black spot disease in pear breeding. Breed Sci 71:240–252
Thomson D, Henry R (1995) Single-step protocol for preparation of plant tissue for analysis by PCR. Biotechniques 19(394–397):400
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40:e115
Van Ooijen JW (2006) JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Van Ooijen JW (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93:343–349
Voorrips RE (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang JP, Tian SL, Sun XL, Cheng XC, Duan NB, Tao JH, Shen GN (2020) Construction of pseudomolecules for the Chinese chestnut (Castanea mollissima) Genome. G3-Genes Genomes Genetics 10:3565–3574
Yamada M, Yamane H, Ukai Y (1994) Genetic-analysis of Japanese persimmon fruit weight. J Am Soc Hortic Sci 119:1298–1302
Yamamoto T, Tanaka T, Kotobuki K, Matsuta N, Suzuki M, Hayashi T (2003) Characterization of simple sequence repeats in Japanese chestnut. J Hortic Sci Biotech 78:197–203
Acknowledgements
We are grateful to Mss. M. Tsukamoto, H. Takahashi, and N. Minagawa for their technical assistance.
Funding
This work was partially supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Innovation, HOR-2004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Data archiving statement
This paper uses existing public data, and there are no new data to register. All of the newly designed primers mapped on linkage map are shown in the table of the manuscript.
Additional information
Communicated by A.M. Dandekar.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Terakami, S., Nishio, S., Kato, H. et al. Fine mapping of the gene controlling the easy-peeling pellicle trait and development of an efficient marker-assisted selection system in Japanese chestnut (Castanea crenata Sieb. et Zucc.). Tree Genetics & Genomes 19, 2 (2023). https://doi.org/10.1007/s11295-022-01575-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01575-6