Abstract
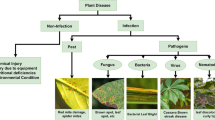
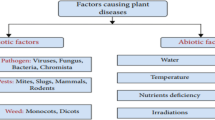
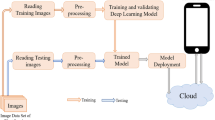
Identification of plant disease is usually done through visual inspection or during laboratory examination which causes delays resulting in yield loss by the time identification is complete. On the other hand, complex deep learning models perform the task with reasonable performance but due to their large size and high computational requirements, they are not suited to mobile and handheld devices. Our proposed approach contributes automated identification of plant diseases which follows a sequence of steps involving pre-processing, segmentation of diseased leaf area, calculation of features based on the Gray-Level Co-occurrence Matrix (GLCM), feature selection and classification. In this study, six color features and twenty-two texture features have been calculated. Support vector machines is used to perform one-vs-one classification of plant disease. The proposed model of disease identification provides an accuracy of 98.79% with a standard deviation of 0.57 on tenfold cross-validation. The accuracy on a self-collected dataset is 82.47% for disease identification and 91.40% for healthy and diseased classification. The reported performance measures are better or comparable to the existing approaches and highest among the feature-based methods, presenting it as the most suitable method to automated leaf-based plant disease identification. This prototype system can be extended by adding more disease categories or targeting specific crop or disease categories.







Similar content being viewed by others
Availability of Data and Material
The data used for experiments is available at https://github.com/spMohanty/PlantVillage-Dataset.
Code Availability
The source code of the project will be made public after acceptance of the manuscript.
References
Savary, S., Ficke, A., Aubertot, J. N., & Hollier, C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Springer.
Pertot, I., Kuflik, T., Gordon, I., Freeman, S., & Elad, Y. (2012). Identificator: A web-based tool for visual plant disease identification, a proof of concept with a case study on strawberry. Computers and Electronics in Agriculture, 84, 144–154.
Colwell, R. (1956). Determining the prevalence of certain cereal crop diseases by means of aerial photography. Hilgardia, 26(5), 223–286.
Mahlein, A.-K. (2016). Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Disease, 100(2), 241–251.
Bock, C. H., & Nutter, F. W. (2011). Detection and measurement of plant disease symptoms using visible-wavelength photography and image analysis. Plant Sciences Reviews, 2012, 73.
Bock, C. H., Poole, G. H., Parker, P. E., & Gottwald, T. R. (2010). Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Critical Reviews in Plant Sciences, 29(2), 59–107.
Shafi, R., Shuai, W., & Younus, M. U. (2020). 360-degree video streaming: A survey of the state of the art. Symmetry, 12(9), 1491.
Shafi, R., Shuai, W., & Younus, M. U. (2020). MTC360: A multi-tiles configuration for viewport-dependent 360-degree video streaming. In 2020 IEEE 6th international conference on computer and communications (ICCC) (pp. 1868–1873). IEEE.
Savary, S., Teng, P. S., Willocquet, L., & Nutter, F. W. (2006). Quantification and modeling of crop losses: A review of purposes. Annual Review of Phytopathology, 44, 89–112.
Dhingra, G., Kumar, V., & Joshi, H. D. (2017). Study of digital image processing techniques for leaf disease detection and classification. Multimedia Tools and Applications, 77, 1–50.
Dhingra, G., Kumar, V., & Joshi, H. D. (2018). Study of digital image processing techniques for leaf disease detection and classification. Multimedia Tools and Applications, 77(15), 19951–20000.
Hughes, D., & Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060.
Barbedo, J. G. A. (2019). Plant disease identification from individual lesions and spots using deep learning. Biosystems Engineering, 180, 96–107.
Ferentinos, K. P. (2018). Deep learning models for plant disease detection and diagnosis. Computers and Electronics in Agriculture, 145, 311–318.
Khandelwal, I., & Raman, S. (2019). Analysis of transfer and residual learning for detecting plant diseases using images of leaves. In Computational intelligence: Theories, applications and future directions (vol. II, pp. 295–306). Springer.
Mohanty, S. P., Hughes, D. P., & Salathé, M. (2016). Using deep learning for image-based plant disease detection. Frontiers in Plant Science, 7, 1419.
Zhang, S., Wang, H., Huang, W., & You, Z. (2018). Plant diseased leaf segmentation and recognition by fusion of superpixel, K-means and PHOG. Optik-International Journal for Light and Electron Optics, 157, 866–872.
Goncharov, P., Ososkov, G., Nechaevskiy, A., & Uzhinskiy, A. (2018). Architecture and basic principles of the multifunctional platform for plant disease detection.
Li, J., Jia, J., & Xu, D. (2018). Unsupervised representation learning of image-based plant disease with deep convolutional generative adversarial networks. In 2018 37th Chinese control conference (CCC). IEEE.
Phadikar, S., Sil, J., & Das, A. K. (2013). Rice diseases classification using feature selection and rule generation techniques. Computers and Electronics in Agriculture, 90, 76–85.
Baquero, D., Molina, J., Gil, R., Bojacá, C., Franco, H., & Gómez, F. (2014). An image retrieval system for tomato disease assessment. In 2014 XIX symposium on image, signal processing and artificial vision (STSIVA). IEEE.
Patil, J. K., & Kumar, R. (2017). Analysis of content based image retrieval for plant leaf diseases using color, shape and texture features. Engineering in Agriculture, Environment and Food, 10(2), 69–78.
Sandika, B., Avil, S., Sanat, S., & Srinivasu, P. (2016). Random forest based classification of diseases in grapes from images captured in uncontrolled environments. In 2016 IEEE 13th international conference on signal processing (ICSP). IEEE.
Oberti, R., Marchi, M., Tirelli, P., Calcante, A., Iriti, M., & Borghese, A. N. (2014). Automatic detection of powdery mildew on grapevine leaves by image analysis: Optimal view-angle range to increase the sensitivity. Computers and Electronics in Agriculture, 104, 1–8.
Sharif, M., Khan, M. A., Iqbal, Z., Azam, M. F., Lali, M. I. U., & Javed, M. Y. (2018). Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Computers and Electronics in Agriculture, 150, 220–234.
Singh, V., & Misra, A. K. (2017). Detection of plant leaf diseases using image segmentation and soft computing techniques. Information Processing in Agriculture, 4(1), 41–49.
Bai, X., Li, X., Fu, Z., Lv, X., & Zhang, L. (2017). A fuzzy clustering segmentation method based on neighborhood grayscale information for defining cucumber leaf spot disease images. Computers and Electronics in Agriculture, 136, 157–165.
Picon, A., Alvarez-Gila, A., Seitz, M., Ortiz-Barredo, A., Echazarra, J., & Johannes, A. (2018). Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Computers and Electronics in Agriculture, 161, 280–290.
Yuan, Y., Fang, S., & Chen, L. (2018). Crop disease image classification based on transfer learning with DCNNs. Springer.
Zeng, W., Li, M., Zhang, J., Chen, L., Fang, S., & Wang, J. (2018). High-order residual convolutional neural network for robust crop disease recognition. In Proceedings of the 2nd international conference on computer science and application engineering. ACM.
Shafarenko, L., Petrou, M., & Kittler, J. (1997). Automatic watershed segmentation of randomly textured color images. IEEE Transactions on Image Processing, 6(11), 1530–1544.
Vala, H. J., & Baxi, A. (2013). A review on Otsu image segmentation algorithm. International Journal of Advanced Research in Computer Engineering & Technology (IJARCET), 2(2), 387–389.
Saleem, G., Akhtar, M., Ahmed, N., & Qureshi, W. S. (2019). Automated analysis of visual leaf shape features for plant classification. Computers and Electronics in Agriculture, 157, 270–280.
Ahmed, N., Khan, U. G., & Asif, S. (2016). An automatic leaf based plant identification system. Science International, 28(1), 427–430.
Clausi, D. A. (2002). An analysis of co-occurrence texture statistics as a function of grey level quantization. Canadian Journal of Remote Sensing, 28(1), 45–62.
Soh, L.-K., & Tsatsoulis, C. (1999). Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Transactions on Geoscience and Remote Sensing, 37, 47.
Haralick, R. M., Shanmugam, K., & Dinstein, I. H. (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics, 3(6), 610–621.
Santos, T. A., Maistro, C. E. B., Silva, C. B., Oliveira, M. S., França, M. C., & Castellano, G. (2015). MRI texture analysis reveals bulbar abnormalities in Friedreich ataxia. American Journal of Neuroradiology, 36, 2214–2218.
Grus, J. (2015). Data science from scratch: First principles with python. O’Reilly Media, Inc.
Liu, H., & Yu, L. (2005). Toward integrating feature selection algorithms for classification and clustering. IEEE Transactions on Knowledge and Data Engineering, 17(4), 491–502.
Islam, M., Dinh, A., Wahid, K., & Bhowmik, P. (2017). Detection of potato diseases using image segmentation and multiclass support vector machine. In 2017 IEEE 30th Canadian conference on electrical and computer engineering (CCECE). IEEE.
Hlaing, C. S., & Zaw, S. M. M. (2017). Plant diseases recognition for smart farming using model-based statistical features. In 2017 IEEE 6th global conference on consumer electronics (GCCE). IEEE.
Hlaing, C. S., & Zaw, S. M. M. (2017). Model-based statistical features for mobile phone image of tomato plant disease classification. In 2017 18th international conference on parallel and distributed computing, applications and technologies (PDCAT). IEEE.
Amara, J., Bouaziz, B., & Algergawy, A. (2017). A deep learning-based approach for banana leaf diseases classification. In BTW (workshops).
Durmuş, H., Güneş, E. O., & Kırcı, M. (2017). Disease detection on the leaves of the tomato plants by using deep learning. In 2017 6th international conference on agro-geoinformatics. IEEE.
Brahimi, M., Boukhalfa, K., & Moussaoui, A. (2017). Deep learning for tomato diseases: Classification and symptoms visualization. Applied Artificial Intelligence, 31(4), 299–315.
Yamamoto, K., Togami, T., & Yamaguchi, N. (2017). Super-resolution of plant disease images for the acceleration of image-based phenotyping and vigor diagnosis in agriculture. Sensors, 17(11), 2557.
Wang, J., Chen, L., Zhang, J., Yuan, Y., Li, M., & Zeng, W. (2018). CNN transfer learning for automatic image-based classification of crop. In Image and graphics technologies and applications: 13th conference on image and graphics technologies and applications, IGTA 2018, Beijing, China, April 8–10, 2018, revised selected papers. Springer.
Yadav, R., Rana, Y. K., & Nagpal, S. (2018). Plant leaf disease detection and classification using particle swarm optimization. Springer.
Kour, V. P., & Arora, S. (2019). Particle swarm optimization based support vector machine (P-SVM) for the segmentation and classification of plants. IEEE Access, 7, 29374–29385.
Pardede, H. F., Suryawati, E., Sustika, R., & Zilvan, V. (2018). Unsupervised convolutional autoencoder-based feature learning for automatic detection of plant diseases. In 2018 international conference on computer, control, informatics and its applications (IC3INA). IEEE.
Suryawati, E., Sustika, R., Yuwana, R. S., Subekti, A., & Pardede, H. F. (2018). Deep structured convolutional neural network for tomato diseases detection. In 2018 international conference on advanced computer science and information systems (ICACSIS). IEEE.
Baranwal, S., Khandelwal, S., & Arora, A. (2019). Deep learning convolutional neural network for apple leaves disease detection. Available at SSRN 3351641.
Funding
There are no funding sources for this Project.
Author information
Authors and Affiliations
Contributions
The contributions of the authors are listed below: NA: Conception, coding, experimentations and writeup. HMSA: Conception, supervision and review. GS: Coding, experimentations, writeup and review. MUY: experimentations, writeup and review. SA: Proof Read and reviewed the manuscript. MRA: Proof Read and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
It is declared that there are no financial and other competing conflicts of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmad, N., Asif, H.M.S., Saleem, G. et al. Leaf Image-Based Plant Disease Identification Using Color and Texture Features. Wireless Pers Commun 121, 1139–1168 (2021). https://doi.org/10.1007/s11277-021-09054-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-021-09054-2