Abstract
Farmers face the formidable challenge of meeting the increasing demands of a rapidly growing global population for agricultural products, while plant diseases continue to wreak havoc on food production. Despite substantial investments in disease management, agriculturists are increasingly turning to advanced technology for more efficient disease control. This paper addresses this critical issue through an exploration of a deep learning-based approach to disease detection. Utilizing an optimized Convolutional Neural Network (E-CNN) architecture, the study concentrates on the early detection of prevalent leaf diseases in Apple, Corn, and Potato crops under various conditions. The research conducts a thorough performance analysis, emphasizing the impact of hyperparameters on plant disease detection across these three distinct crops. Multiple machine learning and pre-trained deep learning models are considered, comparing their performance after fine-tuning their parameters. Additionally, the study investigates the influence of data augmentation on detection accuracy. The experimental results underscore the effectiveness of our fine-tuned enhanced CNN model, achieving an impressive 98.17% accuracy in fungal classes. This research aims to pave the way for more efficient plant disease management and, ultimately, to enhance agricultural productivity in the face of mounting global challenges. To improve accessibility for farmers, the developed model seamlessly integrates with a mobile application, offering immediate results upon image upload or capture. In case of a detected disease, the application provides detailed information on the disease, its causes, and available treatment options.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
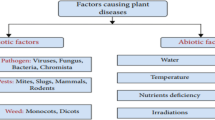
Plant diseases are now a major worldwide issue, causing food shortages in numerous nations. Identifying these diseases accurately is essential for increasing agricultural yields. Convolutional Neural Networks (CNNs) have become increasingly popular in today's artificial intelligence era as efficient instruments for accurate disease identification through picture analysis. As a result, CNNs have become a prevalent choice for the early detection of crop diseases, leading to improved accuracy (Bangari et al. 2022; Singh et al. 2023; Asif et al. 2020), and the expansion of technology's role in precision agriculture. Detecting diseases in maize leaves typically requires continuous crop monitoring. According to professionals, these methods can incur high expenses, consume significant time, and exhibit inconsistencies. To address these challenges, CNNs are employed to automate the detection and classification of maize leaf diseases swiftly and accurately (Camargo and Smith 2009). Numerous researchers have developed predictive models to determine whether maize leaves will succumb to disease or remain healthy (Zhang et al. 2021). Figure 1 likely shows a visual representation distinguishing between plant diseases caused by infections (such as pathogens like bacteria, viruses, fungi, or pests) and diseases resulting from non-infectious factors (such as nutrient deficiencies, environmental stress, or genetic factors).
Enhancing agricultural systems becomes imperative as the global population is projected to reach 10 billion in the next 25 years. Leveraging artificial intelligence, particularly Convolutional Neural Networks (CNNs), can help farmers increase food production, mitigating the looming threat of food scarcity, a potential leading cause of mortality.
CNNs are excellent at object detection and image classification tasks because they can automatically extract complicated hierarchical patterns from unprocessed data. There are many reasons to use the CNN model for detecting plant diseases. CNNs are particularly good at extracting valuable features from photos of leaves or plant tissues, even in the presence of differences caused by different lighting conditions, backdrops, and plant species. This is important for plant disease detection, where identifying subtle visual cues suggestive of diseases is critical. This architecture makes it possible for CNNs to efficiently capture local patterns and spatial relationships, crucial for identifying disease signs that might appear in plant leaf or tissue regions. Various elements, including plant growth stages, weather circumstances, and the presence of weeds or other artifacts, contribute to the inherent diversity of agricultural landscapes. CNNs exhibit strength to such variations, enabling them to generalize well across different conditions and effectively detect diseases in plants regardless of variations in appearance and environmental conditions. While using CNN models, remember that they are lightweight and fast in image recognition in any environmental conditions.
There are still many things that could be improved in the previous work. For instance, K. Sathya (Sathya and Rajalakshmi 2022) proposed their model RDA-CNN, inspired by CNN architecture, achieving an accuracy of 96.59%. However, they did not test their model by integrating it into a mobile application. Similarly, Md. Tariqul Islam (Islam 2020) achieved a 94.29% accuracy with their proposed model, but they also did not create any system to test their model. Shradha Verma (Verma et al. 2020) presented a CNN model by tuning hyperparameters, but their results were not sufficiently accurate, and they did not create a system to test their model either.
One significant development in agriculture is the automated diagnosis of plant diseases using information from plant leaves. Furthermore, crop quality and yield are positively impacted by the on-time and accurate detection of plant diseases. Because of the cultivation of a vast array of crop products, even a pathologist or agriculturist may frequently be unable to recognize plant diseases by observing diseased leaves. Visual inspection is the predominant method of disease identification in rural parts of developing countries. Expert monitoring is also necessary constantly. Farmers in isolated places might have to make a long and costly journey to an expert's consultation. Agronomists and farmers benefit from automated computational systems' high precision and throughput in identifying and diagnosing plant diseases. The suggested technique enables reliable pattern recognition by using CNNs' capacity to extract intricate details from plant photos. The proposed model is more optimized and faster and does accurate prediction and classification with optimized hyperparameters. The Proposed model learns to differentiate between illnesses based on optical indicators using a dataset of healthy and diseased plant samples. Applying machine learning approaches enhances the detection process's efficacy and accuracy and is valuable for crop disease identification. The paper aims to improve precision agriculture by developing a scalable method that will decrease the effect of diseases of plants on crops. As mentioned in Fig. 2 basic architecture of the CNN model. In the end, this paper presents a mobile-based system for detecting leaf disease using (E-CNN); initially, the model was converted into a TF Lite file by integrating the mobile application. The proposed model can feed images directly from farmers' mobile devices. The model then classifies plant diseases and detects objects. It then shows the type of ailment, its confidence percentage, and how long it took to classify the image. We created an Android smartphone app to help farmers with limited resources take pictures of the sick plant leaves. On the user's end, the mobile app operates on top of the CNN paradigm. The application also shows the processed image's categorization time and confidence percentage.
This research focuses on exploring fine tuning and hyperparameter optimization on multiple popular pre-trained CNN based models from literature to address farmers' disease detection and classification concerns. Moreover, we implement the best performing model into a mobile application presenting an easy way to apply the fine-tuned model on different plants and get details about the disease and possible treatment in friendly manner.
This research centers around optimizing disease detection for farmers through Convolutional Neural Networks (CNNs). The study makes four key contributions:
-
Evaluation and comparison of popular pre-trained CNNs based models (e.g., AlexNet (Durmuş et al. 2017), MobileNet (Daphal and Koli 2023; Bi et al. 2022) and ResNet (Nachtigall 2016) with finely tuned hyperparameters in addition to some machine learning models like SVM.
-
Utilization of advanced machine learning models for efficient plant disease detection, providing a sophisticated approach to agricultural disease management.
-
Refinement of CNN hyperparameters, including activation functions, Epochs, learning rate, optimizer, and number of layers, through grid search and proper dataset splitting methods to enhance performance of all the considered models deep learning models.
-
Seamless integration into a user-friendly mobile app for efficient disease detection by capturing and processing real-time images. Detected diseases trigger immediate results with additional information on details, causes, and treatment options.
The rest of the paper is organized as follows. Section 2 presents related works. The formulation of the problem and the fundamental idea underlying the proposed method are detailed in Sect. 3. Section 4 discusses the experimental results and the different outcomes. In addition, it presents the Challenges in applying the proposed plant disease model and its limitations respectively. The work is concluded in Section 5.
2 Related works
Numerous plant diseases and research papers addressing different ailments were previously examined. Our research concentrated on potato, apple, and corn diseases, specifically investigating healthy potato leaves, early blight potato leaves, late blight potato leaves, and diseases of apple leaves and corn. Convolutional Neural Networks (CNNs) were harnessed as a tool for accurate disease detection. In this context, we explored various hyperparameters such as optimizers, activation functions, and dropout rates, seeking optimal configurations that maximize accuracy (Sankaran et al. 2010). It is worth noting that previous research primarily employed Grid Search for hyperparameter optimization, which is an exhaustive search method. Our literature review revealed a wealth of research papers dedicated to plant disease detection using various techniques (Barman et al. 2020; Poojary and Pai 2019; Butale and Kodavade 2019; Saleem et al. 2020). This section of our paper underscored the significance of these valuable studies within the context of CNN models.
The subsequent section overviewed different CNN architectures for plant disease diagnosis. CNNs, typically adopting the Lenet-5 architecture proposed by Yann LeCun in 1998 (Yi et al. 2018), served as the common architecture. Convolution layers were frequently employed for feature extraction, followed by pooling techniques like max, min, or average pooling (Sankaran et al. 2010). The final layer comprised a fully connected layer for disease classification. This study employed CNN-based deep-learning techniques for plant disease identification. The procedure usually comprised gathering high-quality photos of both healthy and diseased leaves for the dataset, which was then divided into training (80%), validation (10%), and testing (10%) groups. For training, 80% of the dataset is enough, while when researchers have to do validation and testing, 10% for each is enough. The use of AlexNet and other deep learning models for predicting leaf illnesses was also covered in the study, along with a description of a novel model for leaf prediction and identification (Liu et al. 2018). The NAG Algorithm was used to optimize these models, which showed remarkable accuracy in recognizing illness traits from pictures.
Additionally, we looked at studies that used optimizers and pre-processing methods in conjunction with CNNs to identify diseases (Subramanian et al. 2022) accurately. The study also investigated feature extraction, picture-enhancing techniques, and classification algorithms such as ANN, SVM, and FUZZY (Pujari et al. 2016). Moreover, researcher developed Hybrid Convolutional Support Machine (H-CSM) combining SVM, CNN and CBAM for detecting disease in plant leaf with complexity pattern and inter class similarity (Kaur, et al. 2024). The selection of image datasets has a significant impact on research outcomes in the field of image analysis currently because image selection is an integral part of model training. So, though their scale and complexity presented processing hurdles, colored images provided a rich visual depiction. Although grayscale images were less dependent on color, they were less computationally efficient. Although segmented images needed labor-intensive annotation, they were beneficial for accurate object location (Sharma et al. 2019). The choice of datasets must be carefully considered by researchers considering their goals for the study, as model training mainly depends on datasets and the harmony between the complexity and richness of the data. Table 1 depicts the dataset used, the ML/DL model, and the detection use cases for recent studies in the field.
In particular, Durmus et al. (Durmuş et al. 2017) used a deep learning method to detect disease in tomato leaves. This method uses two different deep learning models: (i) AlexNet and (ii) SqueezeNet. The training was done with the help of ten different apple, potato, and corn classes, and the accuracy was 95.65% and 94.30% for AlexNet and SqueezeNet, respectively. In (Liu et al. 2018), the authors applied the detection on Apple leaves using CNN's pre-trained AlexNet model. The model is trained to recognize four significant apple leaf diseases, achieving up to 97.62% accuracy. Another study by DeChant et al. (DeChant et al. 2017) conducted automatic identification of the Northern leaf blight of Maize leaf through a computation pipeline of CNN that shows the challenging part of the limited data. The system got accuracy up to 96.7%. Furthermore, AlexNet and GoogLeNet architectures for deep learning were used by Mohanty et al. (Mohanty et al. 2016) to produce models for categorizing tomato leaf diseases. By combining learning methods and different training and testing splits, their system got an accuracy of 99.35% using the PlantVillage (PlantVillage Dataset 2023) dataset.
Ahmed et al. (Ahmad et al. 2020) used four different Pre-trained CNN networks, VGG-16, ResNet50, Inception V3, and VGG-19, for identification and classification of tomato leaf disease, where the model got 93% accuracy. The next study was by Rumpf et al. in 2010, when he discussed the differences between healthy and unhealthy leaves of sugar beet,he trained his model in such a way that without disease particular symptoms visibility on leaves, the model identified the disease by using SVM and their model got the accuracy of 97%.
The detection of soybean diseases was a challenging task due to variations in natural scene backgrounds. To deal with this, researchers have developed an approach utilizing Choquet fuzzy ensemble strategy with CNN for the detection and classification of soybean diseases with high accuracy and precision (Hang et al. 2024; Mishra et al. 2022). Enhanced corn seed disease classification and detection, using feature augmentation and transfer learning, include data augmentation such as rotation, flipping, and scaling are investigated to enhance the robustness of CNN model in detecting skin cancer lesion (Rai and Roop 2024; Patel and Atul 2024). Moreover, to detect leaf illness, this study contrasts a newly created custom CNN with nine pre-trained models. With its efficiency increasing in training speed and memory needs, the custom CNN presents a strong substitute for agricultural leaf disease early detection (Alam et al. 2024).
In (Nachtigall 2016), Nachtigall et al. used CNN in the detection and grouping of leaf disease also, nutritional deficiencies, and herbicide damage on apple leaves. Their system got an accuracy of 97.3%, and the dataset was up to 2539 images. Recently, Swapnil Dadabhau Daphal et al. (Daphal and Koli 2023) collected their dataset of up to 2569 images with five categories. The author used the well-known deep learning technique MobileNet-V2, proposing the system for better generalization, and the system got an accuracy of 84%. The primary contribution of this study lies in the comprehensive analysis of hyperparameters in the context of plant disease detection, emphasizing three specific classes while demonstrating the effectiveness of the fine-tuned model. This research study provides valuable insights by evaluating the different CNN-based networks used in the field, offering a foundation for future research in plant disease detection.
3 Methodology
In this section, the architecture of the CNN model and our proposed model, E-CNN, for detecting and classifying plant diseases is elucidated comprehensively.
3.1 The architecture of CNN
The basic architecture of a Convolutional Neural Network (CNN) is depicted in Fig. 2, representing a fundamental framework in deep learning, particularly for image processing tasks. CNNs are designed to analyze images by assigning adjustable biases and weights to various elements, allowing them to discern different visual aspects. The initial phase of a CNN involves manual filter creation during training, gradually refining them to recognize diverse features within the training data.
The structural blueprint of CNNs closely emulates the neural connectivity patterns observed in the human brain, specifically drawing inspiration from the organization of the Visual Cortex. In this biological analogy, individual neurons respond to specific stimuli within a confined visual field, termed the receptive field. These receptive fields overlap, collectively analyzing the entire visual area. Similarly, CNNs adopt this concept to ensure a comprehensive interpretation of the visual input, enhancing their ability to recognize intricate patterns (Ahmad et al. 2020; Nachtigall 2016; Mishra et al. 2022). This biologically inspired approach contributes to the network's efficiency in image processing and feature extraction tasks.
3.2 Enhanced (E-CNN) model for plant disease detection and classification
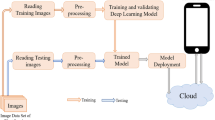
Figure 3 visually represents the entire workflow of our enhanced E-CNN Disease Detection Model. It offers a comprehensive overview of the model's functionality and the distinct stages it undergoes for efficient disease detection. In the next subsections, we delve into a detailed breakdown of the components and experimentation settings.
While Fig. 2 illustrates the structure of the basic CNN, Fig. 4 illustrates the E-CNN model's design that is used by the general Algorithm reported in Algorithm I to classify the output based on a given input image.
In fact, CNNs, inspired by the human visual system, are extensively used in image processing and analysis (Shrestha et al. 2020). Their strength lies in classifying unprocessed input data by autonomously determining the most appropriate filters for this task. This inherent ability of CNNs to extract and recognize features reduces the computational demands typically encountered in other machine-learning methods for these functions (Chauhan et al. 2018). Key elements of a CNN include convolutional, pooling, and fully connected layers, as outlined in summary Table 2.
In contrast, Artificial Neural Networks (ANNs) usually have three distinct layers: Input, Hidden, and Output. Specific bias values and weights characterize neurons in the Hidden layer. These are multiplied with input values and passed through an activation function. If the output value surpasses a certain threshold, it gets forwarded to the next layer in the network; if not, no data is transmitted. ANNs function feed-forwardly, meaning data moves in sequence through the layers. The main goal in adjusting the model is to lower the cost function for each input. The primary objective during model adjustment is minimizing each input's cost function. CNNs represent a neural network with one or more layers designed to extract dependencies from inputs, such as text and images. A key feature of CNNs is the convolution operation performed across multiple intermediate layers. Convolution involves the dot-product of input bundles with a grid structure and a set of weights.
CNNs are particularly popular in image processing and recognition. CNN architectures have seen significant advances, with LeNet-5 1998 being a notable milestone. Earlier computer vision techniques relied on feature identification, demanding substantial expertise in image processing. However, CNNs have transformed image processing by automating feature extraction. They are compatible with matrices, RGB color images, and even tensors, enabling image classification, segmentation, face detection, and object identification. CNNs have found successful applications in diverse fields, including healthcare, web services, mail, and natural language processing. A CNN comprises stacked layers, including convolution, pooling, ReLU activation, and fully connected layers, as mentioned in Fig. 4. These layers process each input image through filtering, correction, and reduction stages before being transformed into a vector. The convolution layer is instrumental in teaching CNNs to recognize specific features, such as object detection. Multiple convolution layers can be employed for added efficiency. The pooling layer further enhances efficiency by down-sampling, significantly reducing computational demands.
A CNN architecture using the Keras Sequential model was designed for image classification. The initial operation, assumed to be rescaling, adjusts pixel values from the typical 0–255 range to a normalized scale of 0–1. The network then sequentially applies Conv2D layers with 32, 64, and 64 filters to detect increasingly complex features in the input images. After each Conv2D layer, MaxPooling2D is employed for spatial down sampling, reducing the spatial dimensions of the feature maps. Dropout layers with a dropout rate of 0.1 (deactivating 10% of neurons) are strategically inserted after each MaxPooling2D layer for regularization to prevent overfitting.
This convolution, pooling, and dropout process is repeated for several layers, progressively extracting detailed hierarchical features from the input images. The final part of the network involves flattening the 2D feature maps into a 1D vector, followed by a densely connected layer with 64 units and ReLu activation. Another dropout layer is applied before the output layer, which consists of 10 units, representing the number of classes in the classification task, and utilizes softmax activation for multi-class classification. The architecture is capped off with a final dropout layer before the output, enhancing model generalization. This CNN structure is well-suited for discerning intricate patterns in image data, contributing to accurate classification into one of the specified classes.
Overall, through systematic hyperparameter tuning, the CNN model was optimized to achieve superior performance in disease detection across a range of crops, thereby contributing to more effective plant disease management in agriculture. Initially, a set of hyperparameters was chosen, including learning rate, batch size, number of layers, filter size, dropout rate, and activation functions. These parameters significantly affect the learning process and the model's ability to extract meaningful features from the input images.
Table 3 shows several vital hyperparameters that are set up to affect the learning process during the training phase of a machine learning model. Training is done for this model throughout 100 epochs, where an epoch is one complete iteration over the dataset. The model's weights are updated using the ADAMAX optimizer, and the loss function selection is set to sparse categorical cross-entropy, which denotes a situation in which the target variable is categorical, and the classes are incompatible. To maximize computing effectiveness and memory utilization, the training data is handled in batches of 32 samples at a time. The dataset's images are standardized to 256 pixels, with three channels signifying the representation of RGB color.
3.3 Image processing and augmentation
In Fig. 5, we show three types of samples used for training and testing. Furthermore, as shown in.
Figure 6, Data augmentation was employed during training, encompassing positional techniques like scaling, cropping, flipping, and rotation. Additionally, brightness, contrast, and saturation adjustments were applied to enhance color quality (Enkvetchakul and Surinta 2022), (Nagaraju et al. 2022). Data augmentation also featured random rotations and distortions, along with horizontal flips. This process generated eight enhanced images for each original image. All original and enhanced images were initially normalized by dividing each pixel's value by 255.
Furthermore, the images were resized to match the input requirements of various model architectures. Due to hardware constraints, the input size for all EfficientNet architecture models was set at 132 × 132, ensuring uniform comparisons among models as done in (Al-Shemarry et al. 2019). Table 4 and Fig. 7 report information about different plant disease types.
3.4 Feature extraction and selection
Feature Extraction: First, basic properties like statistical characteristics of colors (like RGB, HSV, and CIELab, min, max, mean, standard deviation, bias, and kurtosis) will be extracted. Textural and morphological elements like Haralick or Local Binary Patterns can be extracted if necessary (Kaur et al. 2022).
3.4.1 Feature selection
The research study can determine the salient characteristics of all diseases and the salient characteristics of nutritious vegetables with feature selection. This model employed filter techniques like Pearson Correlation Coefficient (PCC), Variance, ANOVA and Principal Component Analysis (PCA) to find hidden patterns in the data.
3.4.2 Pearson correlation coefficient (PCC)
PCC is a statistical method used to measure the linear correlation between two variables x and y. The range for this variable is between + 1 to -1; Positive 1 refers to a positive association between variables, and negative 1 means a negative association between variables.
3.4.3 Variance
Variance measures how sensitive the model is to variation in the training set. It is a metric for how much the model's predictions change depending on the subsets of the training set it is trained on. Moreover, it can be defined as how much the predicted values are away from each other. A high variance model runs the risk of being overfitting because it is excessively complicated and captures both the random noise and the deeper trends in the data. Because the model has remembered the training data instead of generalizing, overfitting happens if a model performs outstandingly on overtrained data but badly on unknown data.
3.4.4 ANOVA
ANOVA refers to the Analysis of Variance. It is used to show the difference between two or more than two means. It generally identifies which features are most important in predicting outputs. It is an essential feature of feature selection.
3.4.5 Principal component analysis (PCA)
PCA is used to overcome overfitting errors in the model through different views; it is a statistical procedure that uses orthogonal alteration that fluctuates a set of correlated variables to set uncorrelated variables.
Statistical Analysis is a pivotal scientific instrument, facilitating the examination and collection of extensive datasets. Its primary purpose is to discern prevalent patterns and trends within large data volumes, converting them into insightful and valuable information.
This model uses only one method of variance analysis (ANOVA). This assists t-student test with the most crucial feature to determine the statistical difference between diseased and healthy leaves.
3.5 Mobile app integration
The combination of TensorFlow Lite (TFLite), Firebase, Flutter, and Dart, presents a comprehensive and robust stack for developing machine learning-powered mobile applications (How to Build Mobile Apps with Flutter 2023). TFLite facilitates the deployment of machine learning models on mobile and embedded devices, ensuring efficient and optimized inference. Firebase integration adds a robust backend to the application, offering features like real-time database, authentication, and cloud functions, enhancing the app's functionality and scalability. Flutter, a UI toolkit developed by Google and Dart as its programming language, provides a cross-platsform framework for building aesthetically pleasing and high-performance mobile applications. Including Dart for background, processing ensures smooth and responsive app behavior even when running tasks in the background. Diagrams, presumably used for system architecture or workflow visualization, contribute to the clarity and understanding of the overall application structure. Figure 8 represents the users’ interaction with our mobile application, which integrates our proposed E-CNN model.
There are two options: one is to instantly capture an image of the leaf, and the second is to upload a captured image from the gallery. After uploading or capturing, the image will pass through our proposed E-CNN model, and the user can see the result label on their screen. Users can also see the details, causes, and treatment of that observed disease. In our experiment, Redmi 13C mobile was used to test this E-CNN-based Mobile App. After testing, it was observed that our application took 25 s on average to detect disease from the image; almost 15 s were required to upload or capture the image, while only 10 s on average were required to process the E-CNN model and show the result. The whole testing used a 4G network. Test results are shown in Fig. 16.
4 Results and discussion
4.1 Implementation of the model
This section outlines the implementation and evaluation of the proposed method, E-CNN, utilizing the Scikit-Learn library and Keras, a high-level API based on TensorFlow Lite, within the Python programming language. The research study employed Jupyter Notebook, an interactive web-based machine learning training and testing platform. The dataset utilized in this experiment was obtained from Kaggle, provided by PlantVillage (PlantVillage Dataset 2023), comprising 54,000 images of various plants. For model training, 10% of the dataset was allocated for validation, 10% for testing, and 80% for training. The training process utilized a batch size of 32 and 100 epochs. Additionally, hyperparameters such as the learning rate were set to 0.0001. During training, the performance of different optimizers, including ADAM, ADAMAX, ADAGRAD, and NADAM, were compared, with ADAMAX selected as the optimizer due to its superior accuracy.
4.2 Dataset
To evaluate the proposed E-CNN model, the dataset used in our experiments is publicly available on the Kaggle site named Plant Village (PlantVillage Dataset 2023), which contained 54,000 images of healthy and infected leaves of various crops. After applying the feature selection techniques explained in Sect. 3.4, Table 5 maps the extracted feature meaning to its real name in the dataset file. As an example, the feature mean_c1_rgb represents the mean of red color in the image. The proposed research study centered on the potato, apple, and corn dataset, consisting of 9175 images divided into the train, test, and validation sets. The potato dataset encompassed three distinct classes: healthy, early blight disease, and late blight disease. Apple and corn are also divided into their respective diseases.
4.3 Evaluation metrics
It has been noted that the PlantVillage dataset was used in most of the research. As a result, the classifications' nature and outcome are extremely similar. Additionally, some researchers have classified bean crop diseases using deep learning algorithms. To prevent findings from repeating, the dataset of leaf disease was used with the help of three deep learning models that utilize different optimization techniques. After the DL model and optimization technique were shown to be the most efficient duo, the collection of photos of bean leaves underwent a varied class illnesses classification. We collected images of diseased leaves from the PlantVillage dataset to distinguish diseases.
where TN and TP are the number of accurate guesses made when the actual class is False or True, respectively. Additionally, the numbers FN and FP indicate how many wrong predictions there were when the actual class was True or false accordingly. These metrics are combined to create a graphic known as the receiver's operating characteristic (ROC) curve. This curve illustrates the trade-off between a model's classification mistakes and FN and FP rates. Furthermore, the AUC graph can be generated using the ROC curve. Specifically, the level of separability is represented by AUC, which ROC leads to a probability curve. AUC indicates a model's capacity to perform well in a classification test.
High separability metrics indicate a superior model with an AUC close to one. Conversely, an inferior separability measure is indicated by an AUC close to zero. If sensitivity and 1-specificity correspond with the probability of TP and FP, respectively, the following estimates the AUC. Table 6 shows the confusion matrix that provides a comprehensive view of a model's performance, allowing for the calculation of the various metrics defined above.
In which
And
where i is used as index.
4.4 Performance analysis
In this section, we initially apply classical machine learning methods, Logistic Regression (LR) and Support Vector Machine (SVM), before delving into various Deep Learning approaches, including CNN-based models, and concluding with the results of our Enhanced CNN (E-CNN).
4.4.1 Performance with LR
Logistic Regression is a statistical method used for binary classification, predicting the probability of an instance belonging to a specific class. On the other hand, SVM seeks to find a hyperplane that maximizes the margin between classes, making it versatile for complex decision boundaries in high-dimensional spaces. Logistic Regression, while simpler and interpretable, is more suitable for linearly separable scenarios. Our problem, being more intricate, demonstrates better performance with SVM, as opposed to Logistic Regression, which yields suboptimal results (82% accuracy) due to its linear nature, as reported in Table 7. We employ k-fold cross-validation with an initial value of 10 for this classification.
4.4.2 Comparisons of CNN based pre-trained models
In this subsection, Table 8 reports the main configuration and accuracy results of five popular pre-trained models, out of the listed works in Table 1, on detecting the Fungal disease using the same dataset we have used and explained earlier. An important observation from Table 8 is that the number of parameters versus the number of layers are playing an important role in the measuring the performance. For instance, the highest accuracy was realized by ResNet50 that has 50 layers and 23 million parameters. DensNet121 has the highest number of layer but a smaller number of parameters.
Figures 9 and 10 illustrate the progression of accuracy and loss functions for five popular models that we have fine-tuned, show consistent improvement with respect to their original performance (as pre-trained models with given parameters values). Also, the figures show the evaluation with an increasing number of training and validation epochs. Figure 11 details the evaluation of our E-CNN model that outperformed all the considered models even after the fine-tuning phase.
In particular, we observe from Fig. 9 that pre-trained DenseNet 121 model that we have enhanced by fine-tuning its hyperparameters has achieved an accuracy of 92.20%. It is worth noting that despite its increased accuracy, the parameter count has been reduced from 4.24 million in the original study to a 3.47 million. In the same direction, the pre-trained VGG-16 model demonstrated an impressive 91% accuracy while utilizing 23 layers with 138 parameters as shown in Fig. 10 (the bottom part) and reported in Table 8. The VGG-16 model after our fine tuning outperformed the original study (Subramanian et al. 2022; Ahmad et al. 2020) in terms of accuracy. Moving on to middle part of Fig. 10, the fine-tuned model based on the pre-trained AlexNet (Soujanya and Jabez 2021; Arya and Singh 2019) achieved an accuracy of 87.56% with just eight (8) layers and a minimal parameter count of 60. Thus, the optimized AlexNet model we considered showed superior accuracy compared to the referenced study. Moreover, as shown in Fig. 10 the upper part, our pre-trained ResNet50 model exhibited remarkable accuracy, reaching an impressive 98%, while boasting a substantial 50-layer depth with a parameter count of only 23.
With respect to the number of Epoch impact on these considered models, we observe from Figs. 9 and 10 that by the 10th epoch, the five CNN-based models achieved an accuracy of approximately 70–78% on the potato dataset (see Figs. 9 and 10). Contrastingly, our E-CNN model (refer to Fig. 11) demonstrated a significantly higher accuracy of around 88% at the same epoch. Even after the 100th epoch, the models found in the literature reached an accuracy range of 93–96%, while our E-CNN model surpassed them with an accuracy of 98.17%.
Figure 12 presents the classification results for different machine and deep learning models on the dataset. The Support Vector Machine (SVM) and ResNet50 achieved an accuracy of 96% and 97.8% respectively. However, our E-CNN model achieved an accuracy of 98.2% outperforming all the other models.
In Table 9 we summarize the comparison of the pre-trained models (after fine-tuning and improvement) for plant disease detection using various metrics such as the number of layers, parameter count, dataset size, accuracy, and the types of diseases considered. The highest accuracy recorded among these studies is 98.17%, achieved by the proposed system for Potato disease detection using a CNN approach on the PlantVillage dataset.
4.4.3 Impact of hyperparameters on E-CNN performance
In this subsection we focus on the selected enhanced E-CNN model that outperforms all the other fine-tuned models. We systematically evaluated the influence of various hyperparameters on the performance of the E-CNN model for disease detection across multiple crops. The study encompassed four prominent activation functions (Softmax, Softplus, ReLU, and Sigmoid), along with key parameters like the optimizer, learning rate, batch size, and layer count. The study also evaluates the model's response to an escalating number of epochs. Initial parameterization included the selection of learning rates, batch sizes, layer counts, filter sizes, dropout rates, and activation functions, all crucial factors affecting the learning process and feature extraction capabilities.
Figure 13 depicts the evolution of validation accuracy concerning the Epoch number across different activation functions applied to the E-CNN model. Notably, for a small number of epochs, the Softmax function outperforms the others. However, as the number of epochs increases substantially, all functions demonstrate similar performance, except for Sigmoid, which lags behind in effectiveness even with an extended training duration.
The E-CNN model exhibited promising performance in identifying fungal diseases across various classes, excelling in prevalent diseases like Potato Late Blight and Potato Early Blight. Challenges emerged in recognizing fewer common diseases, resulting in diminished precision and recall for those classes. Fine-tuning hyperparameters, including optimizers, dropouts, and activation functions, revealed unique behaviors, such as challenges faced by the Adagrad optimizer and the Sigmoid activation function. Table 10 reports a detailed analysis that highlights the profound impact of these hyperparameters on model performance. Notably, the highest accuracy observed is 88%, achieved with the "0.1" dropout rate using the Adagrad optimizer. Conversely, the lowest accuracy, 1%, was obtained with the Sigmoid activation function. Figure 14 reports the confusion matrix for the detection of diseases for the three apple, corns and potato plants.
This comparison serves as a reference point to evaluate the performance of the proposed system and its significance in crop disease detection.
Last, Fig. 15 shows the other performance metrics of our proposed improved E-CNN model with the best hyperparameters setup, which has Epoch 100, Optimizer 'Adamax,' Activation Function 'Softmax,' and Dropout ‘0.1’. In our experiment, Redmi 13C mobile was used to test this E-CNN-based Mobile App as shown in Fig. 16.
4.5 Challenges and limitations
There are many challenges and issues involved in disease symptom analysis and developing a CNN model. The issues related to the automatic identification and classification of plant diseases with images is discussed in this section. The challenges conventional plant disease diagnosis systems face are discussed in (Liu et al. 2018). Agricultural environments are subject to various environmental factors, such as lighting conditions, weather fluctuations, and soil composition, which can affect the appearance of plant diseases. Adapting CNN models for environmental variability ensures their effectiveness in real-world settings. This can involve collecting data under diverse environmental conditions, augmenting datasets with synthetic variations, and incorporating domain adaptation techniques into the model training pipeline. Deploying CNN (Khalid and Karan 2023; Nagaraju et al. 2022; Mishra et al. 2022) models in resource-constrained agricultural settings, such as remote farms or developing regions, poses computational resources, power availability, and internet connectivity challenges. To overcome these challenges, researchers develop lightweight CNN architectures optimized for edge devices, implement efficient model inference algorithms, and explore offline processing and data compression techniques to reduce computational overhead.
Also, there are many reasons for developing plant diseases, such as fungi, viruses, and bacteria. Anyone can affect each crop; that is why there would be different classes of crops. Detecting these classes and having specialized knowledge about the disease spectrum for each crop is a must. Diseases within the same class may exhibit disparate indicators across different crops. For example, early blight has small patches on potato leaves; the same is true for tomato leaves. The only difference is the shape of the leaf, which is the same as apple leaves. Powdery mildew disease resembles white powdery patches, while circular lesions with white powdery patches on Potato leaves. Accurately identifying these symptom variations is essential for effective disease detection.
4.6 Limitations of the proposed model.
The proposed E-CNN model faces obstacles and issues in the training and developing intelligent plant disease detection and classification. As any CNN model, the suggested agricultural system is subject to an array of environmental variables, including variations in lighting, weather patterns, and soil composition, all of which influence the appearance of plant diseases. While the proposed model exhibits resilience to certain levels of variability, its ability to generalize effectively across vastly diverse environmental conditions, which is not effectively represented in the training data, may be compromised CNN, especially those using large architectures or needing extensive training data. It should fulfill good hardware specifications and computational requirements. Training models in resource-limited environments, such as low-specification devices or field-based systems, can create hurdles. Addressing these challenges offers detailed features that show the suitability of the proposed CNN-based approach for plant disease detection. Additionally, future research initiatives should be aimed at modifying these constraints and obstacles to advance agricultural disease detection.
5 Conclusion
This paper introduces a novel approach for plant disease detection and classification, leveraging an optimized Convolutional Neural Network (E-CNN) architecture. The study focuses on addressing the pressing challenge faced by farmers in meeting the demands of a rapidly growing global population for agricultural products, amidst the persistent threat of plant diseases. By concentrating on the early detection of prevalent leaf diseases in Apple, Corn, and Potato crops, our research contributes to more efficient disease control and enhanced agricultural productivity. The core CNN structure underwent significant modifications, including the addition of extra network layers, incorporation of a Global Average Pooling (GAP) layer, and integration of a Batch Normalization (BN) layer. Through these structural adjustments, the proposed E-CNN model achieved superior classification accuracy compared to contemporary image classification techniques while reducing the number of parameters. Moreover, the research investigates the influence of data augmentation on detection accuracy, contributing to the reduction of potential overfitting and improving generalization capabilities. To enhance accessibility for farmers, the developed model seamlessly integrates with a mobile application, providing immediate results upon image upload or capture. In case of a detected disease, the application offers detailed information on the disease, its causes, and available treatment options. The experimental results and integration with practical applications underscore the efficacy of our approach in addressing the critical issue of plant disease management. In particular, multiple machine learning and pre-trained deep learning models were considered and fine-tuned, highlighting the effectiveness of our fine-tuned enhanced CNN model with an impressive 98.17% accuracy in fungal classes. The study includes a thorough performance analysis of several models, emphasizing the impact of hyperparameters on plant disease detection across these three distinct crops.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- CNN:
-
Convolutional Neural Network
- E-CNN:
-
Enhanced Convolutional Neural Network
- KNN:
-
K-Nearest Neighbors
- SVM:
-
Support Vector Machine
- ANN:
-
Artificial Neural Network
- SVM:
-
Support Vector Machine
- ML:
-
Machine Learning
- DL:
-
Deep Learning
- TF:
-
Lite Tensor Flow Lite
References
Adedoja A, Owolawi PA, Mapayi T (2019) Deep learning based on nasnet for plant disease recognition using leave images. In: 2019 international conference on advances in big data, computing and data communication systems (icABCD). IEEE, pp 1–5
Ahmad I, Hamid M, Yousaf S, Shah ST, Ahmad MO (2020) Optimizing pretrained convolutional neural networks for tomato leaf disease detection. Complexity 2020:1–6
Alam TS, Chandni BJ, Abhijit P (2024) Comparing pre-trained models for efficient leaf disease detection: a study on custom CNN. J Electr Syst Inf Technol 111:12
Al-Shemarry MS, Li Y, Abdulla S (2019) An efficient texture descriptor for the detection of license plates from vehicle images in difficult conditions. IEEE Trans Intell Transp Syst 21(2):553–564
Arya S, Singh R (2019) A comparative study of cnn and alexnet for detection of disease in potato and mango leaf. In: 2019 International conference on issues and challenges in intelligent computing techniques (ICICT). IEEE, vol 1, pp 1–6
Asif MKR, Rahman MA, Hena MH (2020) CNN based disease detection approach on potato leaves. In: 2020 3rd International conference on intelligent sustainable systems (ICISS). IEEE, pp 428–432
Atila Ü, Uçar M, Akyol K, Uçar E (2021) Plant leaf disease classification using EfficientNet deep learning model. Eco Inform 61:101182
Atole RR, Park D (2018) A multiclass deep convolutional neural network classifier for detection of common rice plant anomalies. Int J Adv Comput Sci Appl 9(1)
Bangari S, Rachana P, Gupta N, Sudi PS, Baniya KK (2022) A survey on disease detection of a potato leaf using cnn. In: 2022 Second international conference on artificial intelligence and smart energy (ICAIS). IEEE, pp 144–149
Barbedo JGA, Koenigkan LV, Santos TT (2016) Identifying multiple plant diseases using digital image processing. Biosys Eng 147:104–116
Barman U, Sahu D, Barman GG, Das J (2020) Comparative assessment of deep learning to detect the leaf diseases of potato based on data augmentation. In: 2020 International conference on computational performance evaluation (ComPE). IEEE, pp 682–687
Bi C, Wang J, Duan Y, Fu B, Kang JR, Shi Y (2022) MobileNet based apple leaf diseases identification. Mobile Networks Appl 27:172–180. https://doi.org/10.1007/s11036-020-01640-1
Butale NM, Kodavade DV (2019) Detection of plant leaf diseases using image processing and soft-computing techniques. Int Res J Eng Technol 6:3288–3291
Camargo A, Smith JS (2009) Image pattern classification for the identification of disease causing agents in plants. Comput Electron Agric 66(2):121–125
Chauhan R, Ghanshala KK, Joshi RC (2018) Convolutional neural network (CNN) for image detection and recognition. In: 2018 First international conference on secure cyber computing and communication (ICSCCC). IEEE, pp 278–282
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA (2020) Using deep transfer learning for image-based plant disease identification. Comput Electron Agric 173:105393
Daphal SD, Koli SM (2023) Enhancing sugarcane disease classification with ensemble deep learning: a comparative study with transfer learning techniques. Heliyon 9(8):e18261. https://doi.org/10.1016/j.heliyon.2023.e18261
PlantVillage Dataset (2023) [Online]. Available: https://www.kaggle.com/datasets/abdallahalidev/plantvillage-dataset. Accessed 24 May 2024
DeChant C, Wiesner-Hanks T, Chen S, Stewart EL, Yosinski J, Gore MA, Lipson H (2017) Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology 107(11):1426–1432
Devi RS, Kumar VR, Sivakumar P (2023) EfficientNetV2 model for plant disease classification and pest recognition. Comput Syst Sci Eng 45(2):2249–2263. https://doi.org/10.32604/csse.2023.032231
Durmuş H, Güneş EO, Kırcı M (2017) Disease detection on the leaves of the tomato plants by using deep learning. In: 2017 6th International conference on agro-geoinformatics. IEEE, pp 1–5
Enkvetchakul P, Surinta O (2022) Effective data augmentation and training techniques for improving deep learning in plant leaf disease recognition. App Sci Eng Prog 15(3):3810–3810
Fuentes A, Yoon S, Kim SC, Park DS (2017) A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 17(9):2022
Hang Y, Xiangyan M, Qiufeng Wu (2024) Application of improved lightweight network and Choquet fuzzy ensemble technology for soybean disease identification. IEEE Access, 12:25146–25163. https://doi.org/10.1109/ACCESS.2024.3365829
Hlaing CS, Zaw SMM (2017) Model-based statistical features for mobile phone image of tomato plant disease classification. In 2017 18th international conference on parallel and distributed computing, applications and technologies (PDCAT). IEEE, pp 223–229
Hlaing CS, Zaw SMM (2018) Tomato plant diseases classification using statistical texture feature and color feature. In: 2018 IEEE/ACIS 17th International conference on computer and information science (ICIS). IEEE, pp 439–444
How to Build Mobile Apps with Flutter (2023) [Online]. Available: https://www.freecodecamp.org/news/how-to-build-mobile-apps-with-flutter/. Accessed 24 May 2024
Hu G, Yang X, Zhang Y, Wan M (2019) Identification of tea leaf diseases by using an improved deep convolutional neural network. Sustain Comput: Inf Syst 24:100353
Islam MT (2020) Plant disease detection using CNN model and image processing. Int J Eng Res Technol 9(10):291–297
Kaur I, Aggarwal G, Verma A (2016) Detection and classification of disease affected region of plant leaves using image processing. Indian J Sci Technol 9:48
Kaur P et al (2022) Recognition of leaf disease using hybrid convolutional neural network by applying feature reduction. Sensors 22.2:575
Kaur P, Mishra AM, Goyal N, Gupta SK, Shankar A, Viriyasitavat W (2024) A novel hybrid CNN methodology for automated leaf disease detection and classification. Expert Syst e13543. https://doi.org/10.1111/exsy.13543
Khalid MM, Karan O (2023) Deep Learning for Plant Disease Detection. Int J Mathem Stat Comput Sci 2:75–84. https://doi.org/10.59543/ijmscs.v2i.8343
Li Y, Nie J, Chao X (2020) Do we really need deep CNN for plant diseases identification? Comput Electron Agric 178:105803
Liu B, Zhang Y, He D, Li Y (2018) Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 10(1):1. https://doi.org/10.3390/sym10010011
Lu Y, Yi S, Zeng N, Liu Y, Zhang Y (2017) Identification of rice diseases using deep convolutional neural networks. Neurocomputing 267:378–384
Mishra AM et al (2022) Weed density estimation in soya bean crop using deep convolutional neural networks in smart agriculture. J Plant Dis Prot 1293:593–604
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419
Nachtigall LG (2016) Classificação de distúrbios em folhas de macieiras utilizando redes neurais convolucionais (Master's thesis, Universidade Federal de Pelotas)
Nagaraju M et al (2022) Convolution network model based leaf disease detection using augmentation techniques. Exp Syst 394:e12885
Ozguven MM, Adem K (2019) Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Physica A 535:122537
Padol PB, Yadav AA (2016) SVM classifier based grape leaf disease detection. In: 2016 Conference on advances in signal processing (CASP). IEEE, pp 175–179
Patel K, Atul P (2024) LeafGuard: efficient soybean leaf defect classification in indian agriculture using fine-tuned CNN. Int J Intell Syst Appl Eng 12.16:16–24
Poojary R, Pai A (2019) Comparative study of model optimization techniques in fine-tuned CNN models. In: 2019 International conference on electrical and computing technologies and applications (ICECTA). IEEE, pp 1–4
Pujari D, Yakkundimath R, Byadgi AS (2016) SVM and ANN based classification of plant diseases using feature reduction technique. IJIMAI 3(7):6–14
Rai CK, Roop P (2024) Northern maize leaf blight disease detection and segmentation using deep convolution neural networks. Multimed Tools Appl 83.7:19415–19432
Ramcharan A, McCloskey P, Baranowski K, Mbilinyi N, Mrisho L, Ndalahwa M, Hughes DP (2019) A mobile-based deep learning model for cassava disease diagnosis. Front Plant Sci 10:425916
Saleem MH, Potgieter J, Arif KM (2020) Plant disease classification: A comparative evaluation of convolutional neural networks and deep learning optimizers. Plants 9(10):1319
Sankaran S, Mishra A, Ehsani R, Davis C (2010) A review of advanced techniques for detecting plant diseases. Comput Electron Agric 72(1):1–13
Sathya K, Rajalakshmi M (2022) RDA-CNN: Enhanced super resolution method for rice plant disease classification. Comput Syst Sci Eng 42(1):33–47. https://doi.org/10.32604/csse.2022.022206
Sharma P, Berwal YPS, Ghai W (2019) Performance analysis of deep learning CNN models for disease detection in plants using image segmentation. Information Processing in Agriculture 7(4):566–574
Shrestha G, Das M, Dey N (2020) Plant disease detection using CNN. In: 2020 IEEE Applied signal processing conference (ASPCON). IEEE, pp 109–113
Singh V, Chug A, Singh AP (2023) Classification of Beans Leaf Diseases using Fine Tuned CNN Model. Proc Comput Sci 218:348–356
Soujanya K, Jabez J (2021) Recognition of plant diseases by leaf image classification based on Improved AlexNet. In: 2021 2nd International conference on smart electronics and communication (ICOSEC). IEEE, pp 1306–1313
Subramanian M, Lv NP, Ve S (2022) Hyperparameter optimization for transfer learning of VGG16 for disease identification in corn leaves using Bayesian optimization. Big Data 10(3):215–229
Tuncer A (2021) Cost-optimized hybrid convolutional neural networks for detection of plant leaf diseases. J Ambient Intell Humaniz Comput 12(8):8625–8636
Verma S, Chug A, Singh AP (2018) Prediction models for identification and diagnosis of tomato plant diseases. In: 2018 International conference on advances in computing, communications and informatics (ICACCI). IEEE, pp 1557–1563
Verma S, Chug A, Singh AP (2020) Impact of hyperparameter tuning on deep learning based estimation of disease severity in grape plant. In: Recent advances on soft computing and data mining: proceedings of the fourth international conference on soft computing and data mining (SCDM 2020). Melaka, Malaysia, January 22– 23, 2020. Springer International Publishing, pp 161–171
Yi N, Li C, Feng X, Shi M (2018) Research and improvement of convolutional neural network. In: 2018 IEEE/ACIS 17th International conference on computer and information science (ICIS). IEEE, pp 637–640
Zhang Y, Wa S, Liu Y, Zhou X, Sun P, Ma Q (2021) High-accuracy detection of maize leaf diseases CNN based on multi-pathway activation function module. Remote Sensing 13(21):4218
Funding
Open Access funding provided by the Qatar National Library. Open Access funding provided by the Qatar National Library.
Author information
Authors and Affiliations
Contributions
I.A.K, M.U and A.D defined the methodology of the work. N.K and A.K enriched the discussion arround the pre-trained DL models and who to fine tune models. M.I developed the mobile application, trained the models and collected the results. M.I and I.A.K produced the figures, M.U and A.D prepared the structure of the tables. All the authors participated in reading and overviewing the state of art and fill accordingly the tables. All the authors contributed in writing some sections and reviewing the sections of the others and discuss the results with respect to the related works.
Corresponding author
Ethics declarations
Ethical approval
This paper does not contain any studies with human participants or animals performed by any authors.
Informed Consent
Informed consent was obtained from all participants in the study.
Human and animal rights
This paper contains no studies with human participants or animals performed by authors.
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iftikhar, M., Kandhro, I.A., Kausar, N. et al. Plant disease management: a fine-tuned enhanced CNN approach with mobile app integration for early detection and classification. Artif Intell Rev 57, 167 (2024). https://doi.org/10.1007/s10462-024-10809-z
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-024-10809-z