Abstract
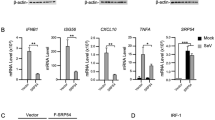
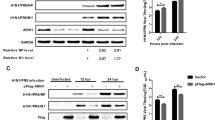
Mitochondrial virus-induced signal adaptor (MAVS), also known as VISA, IPS-1, and Cardif, is a crucial adaptor protein in the RIG-I-like receptor (RLR) signaling pathway. Upon viral infection, RIG-I recognizes viral dsRNA and further transfers it to mitochondria, where it binds to MAVS through its CARD domain, generating a series of signal cascades. Transduction through this signaling cascade leads to phosphorylation and nuclear translocation of interferon regulatory factor 3/7 (IRF3/IRF7) and activation of NF-κB, which ultimately produces type I interferon (IFN) and proinflammatory cytokines. Here, our experiments demonstrated that overexpression of SRY-related high-mobility group protein 9 (SOX9) significantly inhibited Sendai virus (SeV)-induced and MAVS-mediated activation of the IFN-β promoter and ISRE. However, knocking out the expression of SOX9 in cells promoted SeV-induced IFN-β promoter and ISRE activation. Further studies have shown that SOX9 interacts with MAVS and targets MAVS to inhibit the association of MAVS-TRAF2, thereby inhibiting MAVS-mediated TRAF2 ubiquitination. Taken together, these results indicate that SOX9 downregulates IFN-β expression and antiviral signal transduction by targeting MAVS.





Similar content being viewed by others
References
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124(4):783–801. https://doi.org/10.1016/j.cell.2006.02.015
Tan X et al (2018) Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol 72:447–478. https://doi.org/10.1146/annurev-micro-102215-095605
Chow KT, Gale M Jr, Loo YM (2018) RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 36:667–694. https://doi.org/10.1146/annurev-immunol-042617-053309
Berke IC, Li Y, Modis Y (2013) Structural basis of innate immune recognition of viral RNA. Cell Microbiol 15(3):386–394. https://doi.org/10.1111/cmi.12061
Hu MM, Shu HB (2018) Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu Rev Cell Dev Biol 34:357–379. https://doi.org/10.1146/annurev-cellbio-100617-062903
Satoh T et al (2010) LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 107(4):1512–1517. https://doi.org/10.1073/pnas.0912986107
Venkataraman T et al (2007) Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178(10):6444–6455. https://doi.org/10.4049/jimmunol.178.10.6444
Wu B, Hur S (2015) How RIG-I like receptors activate MAVS. Curr Opin Virol 12:91–98. https://doi.org/10.1016/j.coviro.2015.04.004
Hu MM et al (2017) Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med 214(4):973–989. https://doi.org/10.1084/jem.20161015
Yang Q, Shu HB (2020) Deciphering the pathways to antiviral innate immunity and inflammation. Adv Immunol 145:1–36. https://doi.org/10.1016/bs.ai.2019.11.001
Gack MU et al (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446(7138):916–920. https://doi.org/10.1038/nature05732
Oshiumi H et al (2009) Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem 284(2):807–817. https://doi.org/10.1074/jbc.M804259200
Yan J et al (2014) TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol 6(2):154–163. https://doi.org/10.1093/jmcb/mju005
Kell AM, Gale M Jr (2015) RIG-I in RNA virus recognition. Virology 479–480:110–121. https://doi.org/10.1016/j.virol.2015.02.017
Reikine S, Nguyen JB, Modis Y (2014) Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol 5:342. https://doi.org/10.3389/fimmu.2014.00342
Xu LG et al (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19(6):727–740. https://doi.org/10.1016/j.molcel.2005.08.014
Xu LG et al (2012) VISA is required for B cell expression of TLR7. J Immunol 188(1):248–258. https://doi.org/10.4049/jimmunol.1100918
Hou F et al (2011) MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146(3):448–461. https://doi.org/10.1016/j.cell.2011.06.041
Lin CH et al (2001) A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies. Mol Cell 8(3):581–590. https://doi.org/10.1016/s1097-2765(01)00333-1
Fang R et al (2017) MAVS activates TBK1 and IKKepsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog 13(11):e1006720. https://doi.org/10.1371/journal.ppat.1006720
Saha SK et al (2006) Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 25(14):3257–3263. https://doi.org/10.1038/sj.emboj.7601220
Liu S et al (2013) MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. https://doi.org/10.7554/eLife.00785
Pineda G, Ea CK, Chen ZJ (2007) Ubiquitination and TRAF signaling. Adv Exp Med Biol 597:80–92. https://doi.org/10.1007/978-0-387-70630-6_7
Liu B, Gao C (2018) Regulation of MAVS activation through post-translational modifications. Curr Opin Immunol 50:75–81. https://doi.org/10.1016/j.coi.2017.12.002
Xie T et al (2019) RACK1 attenuates RLR antiviral signaling by targeting VISA-TRAF complexes. Biochem Biophys Res Commun 508(3):667–674. https://doi.org/10.1016/j.bbrc.2018.11.203
Li J et al (2020) SNX5 inhibits RLR-mediated antiviral signaling by targeting RIG-I-VISA signalosome. Biochem Biophys Res Commun 522(4):889–896. https://doi.org/10.1016/j.bbrc.2019.11.121
Ling T et al (2018) TARBP2 negatively regulates IFN-beta production and innate antiviral response by targeting MAVS. Mol Immunol 104:1–10. https://doi.org/10.1016/j.molimm.2018.10.017
He TS et al (2018) HAUS8 regulates RLRVISA antiviral signaling positively by targeting VISA. Mol Med Rep 18(2):2458–2466. https://doi.org/10.3892/mmr.2018.9171
Ran FA et al (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308. https://doi.org/10.1038/nprot.2013.143
Joung J et al (2017) Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12(4):828–863. https://doi.org/10.1038/nprot.2017.016
Shalem O et al (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343(6166):84–87. https://doi.org/10.1126/science.1247005
Yang H et al (2020) SOX9 represses hepatitis B virus replication through binding to HBV EnhII/Cp and inhibiting the promoter activity. Antiviral Res 177:104761. https://doi.org/10.1016/j.antiviral.2020.104761
Zhu W et al (2019) TRAF3IP3 mediates the recruitment of TRAF3 to MAVS for antiviral innate immunity. EMBO. https://doi.org/10.15252/embj.2019102075
Jana S et al (2020) SOX9: The master regulator of cell fate in breast cancer. Biochem Pharmacol 174:113789. https://doi.org/10.1016/j.bcp.2019.113789
Panda M, Tripathi SK, Biswal BK (2021) SOX9: An emerging driving factor from cancer progression to drug resistance. Biochim Biophys Acta Rev Cancer 1875(2):188517. https://doi.org/10.1016/j.bbcan.2021.188517
Ren Z et al (2020) Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol 11:1030. https://doi.org/10.3389/fimmu.2020.01030
Chen T et al (2018) Sec13 is a positive regulator of VISA-mediated antiviral signaling. Virus Genes 54(4):514–526. https://doi.org/10.1007/s11262-018-1581-0
Wagner T et al (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79(6):1111–1120. https://doi.org/10.1016/0092-8674(94)90041-8
Chen H et al (2017) Expression and therapeutic potential of SOX9 in chordoma. Clin Cancer Res 23(17):5176–5186. https://doi.org/10.1158/1078-0432.CCR-17-0177
Yuan X et al (2018) SOX9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance. Br J Cancer 119(11):1358–1366. https://doi.org/10.1038/s41416-018-0338-9
Oshima M et al (2018) Virus-like infection induces human beta cell dedifferentiation JCI. Insight. https://doi.org/10.1172/jci.insight.97732
Aleman A et al (2008) Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br J Cancer 98(2):466–473. https://doi.org/10.1038/sj.bjc.6604143
Cheng PF et al (2015) Methylation-dependent SOX9 expression mediates invasion in human melanoma cells and is a negative prognostic factor in advanced melanoma. Genome Biol 16:42. https://doi.org/10.1186/s13059-015-0594-4
Passeron T et al (2009) Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Invest 119(4):954–963. https://doi.org/10.1172/JCI34015
Wang HY, Lian P, Zheng PS (2015) SOX9, a potential tumor suppressor in cervical cancer, transactivates p21WAF1/CIP1 and suppresses cervical tumor growth. Oncotarget 6(24):20711–22. https://doi.org/10.18632/oncotarget.4133
Acknowledgements
We are grateful to Dr. Hong-Bing Shu (Medical Research Institute, Wuhan University) for providing plasmids and other reagents. This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81971502, 82060298).
Author information
Authors and Affiliations
Contributions
LX designed the research. XJ performed the experiments. LX and XJ did data analysis and discussion. XJ and LX wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Joachim Jakob Bugert.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, X., Xu, LG. SOX9 negatively regulates the RLR antiviral signaling by targeting MAVS. Virus Genes 58, 122–132 (2022). https://doi.org/10.1007/s11262-022-01886-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-022-01886-9