Abstract
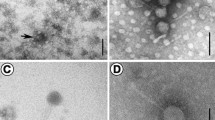
A novel Vibrio phage, P23, belonging to the family Siphoviridae was isolated from the surface water of the Yellow Sea, China. The complete genome of this phage was determined. A one-step growth curve showed that the latent period was approximately 30 min, the burst size was 24 PFU/cell, and the rise period was 20 min. The phage is host specific and is stable over a range of pH (5–10) and temperatures (4–65 °C). Transmission electron microscopy showed that phage P23 can be categorized into the Siphoviridae family, with an icosahedral head of 60 nm and a long noncontractile tail of 144 nm. The genome consisted of a linear, double-stranded 40.063 kb DNA molecule with 42.5% G+C content and 72 putative open reading frames (ORFs) without tRNA. The predicted ORFs were classified into six functional groups, including DNA replication, regulation and nucleotide metabolism, transcription, phage packaging, phage structure, lysis, and hypothetical proteins. The Vibrio phage P23 genome is a new marine Siphoviridae-family phage genome that provides basic information for further molecular research on interaction mechanisms between bacteriophages and their hosts.





Similar content being viewed by others
References
Grimes DJ, Johnson CN, Dillon KS, Flowers AR, Iii NFN, Berutti T (2009) What genomic sequence information has revealed about Vibrio ecology in the ocean—a review. Microb Ecol 58:447–460. https://doi.org/10.1007/s00248-009-9578-9
Almasi A (2005) An investigation on pathogenic Vibrios distribution in domestic wastewater. Iran J Environ Health Sci Eng 2(3):153–157
Almeida A, Cunha A, Gomes NCM, Alves E, Costa L, Faustino MAF (2009) Phage therapy and photodynamic therapy: low environmental impact approaches to inactivate microorganisms in fish farming plants. Marine Drugs 7:268–313. https://doi.org/10.3390/md7030268
Yaashikaa PR, Saravanan A, Kumar PS (2016) Isolation and identification of Vibrio cholerae and Vibrio parahaemolyticus from prawn (Penaeus monodon) seafood: preservation strategies. Microb Pathog 99:5–13. https://doi.org/10.1016/j.micpath.2016.07.014
Fuhrman JA, Suttle CA (1993) Viruses in marine planktonic systems. Oceanography 6(2):51–63. https://doi.org/10.5670/oceanog.1993.14
Suttle CA (1994) The significance of viruses to mortality in aquatic microbial communities. Microb Ecol 28(2):237–243. https://doi.org/10.1007/bf00166813
Malki K, Kula A, Bruder K, Sible E, Hatzopoulos T, Steidel S, Watkins SC, Putonti C (2015) Bacteriophages isolated from Lake Michigan demonstrate broad host-range across several bacterial phyla. Virol J 12:164. https://doi.org/10.1186/s12985-015-0395-0
Ghai R, Martin-Cuadrado AB, Motto AG, Heredia IG, Cabrera R, Martin J, Verdú M, Deschamps P, Moreira D, López-García P, Mira A, Rodriguez-Valera F (2010) Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4:1154–1166. https://doi.org/10.1038/ismej.2010.44
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC (2007) The sorcerer II Global Ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. https://doi.org/10.1371/journal.pbio.0050077
Duhaime MB, Wichels A, Waldmann J, Teeling H, Glöckner FO (2011) Ecogenomics and genome landscapes of marine Pseudoalteromonas phage H105/1. ISME J Multidiscip J Microb Ecol 5:107–121. https://doi.org/10.1038/ismej.2010.94
Li Y, Wang M, Liu Q, Song X, Wang D, Ma Y, Jiang Y (2016) Complete genomic sequence of bacteriophage H188: a novel Vibrio kanaloae phage isolated from Yellow Sea. Curr Microbiol 72:628–633. https://doi.org/10.1007/s00284-015-0984-6
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218. https://doi.org/10.1016/0076-6879(63)06240-6
Hyman P, Abedon ST (2010) Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. https://doi.org/10.1016/s0065-2164(10)70007-1
Liu ZY, Li HF, Wang M, Jiang Y, Yang QW, Zhou XH, Gong Z, Liu Q, Shao HB (2018) Isolation, characterization and genome sequencing of the novel phage SL25 from the Yellow Sea, China. Marine Genom 37:31–34. https://doi.org/10.1016/j.margen.2017.09.008
Ghosh K, Senevirathne A, Kang HS, Hyun WB, Kim JE, Kim KP (2018) Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting bacteriophage BSP10 and its effect on poly- gamma-glutamic acid degradation. Viruses 10:240. https://doi.org/10.3390/v10050240
Pajunen M, Kiljunen S, Skurnik M (2000) Bacteriophage φYeO3-12, Specific for Yersinia enterocolitica Serotype O:3, is related to coliphages T3 and T7. J Bacteriol 182:5114–5120. https://doi.org/10.1128/JB.182.18.5114-5120.2000
Gong Z, Wang M, Yang QW, Li ZS, Xia J, Gao Y et al (2017) Isolation and complete genome sequence of a novel Pseudoalteromonas phage PH357 from the Yangtze River estuary. Curr Microbiol 74:1–8. https://doi.org/10.1007/s00284-017-1244-8
Capra ML, Quiberoni A, Reinheimer JA (2004) Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol 38:499–504. https://doi.org/10.1111/j.1472-765X.2004.01525.x
Alonso MDC, Rodríguez J, Borrego JJ (2002) Characterization of marine bacteriophages isolated from the Alboran Sea (Western Mediterranean). J Plankton Res 24(10):1079–1087. https://doi.org/10.1093/plankt/24.10.1079
Alagappan KM et al (2010) Occurrence of Vibrio parahaemolyticus and its specific phages from shrimp ponds in East Coast of India. Curr Microbiol 61:235–240. https://doi.org/10.1007/s00284-010-9599-0
DrulisKawa Z, Mackiewicz P, KesikSzeloch A et al (2011) Isolation and characterisation of KP34-a novel uKMV-like bacteriophage for Klebsiella pneumoniae. Appl Microbiol Biotechnol 90:1333–1345. https://doi.org/10.1007/s00253-011-3149-y
Othman BA, Askora A, Abo-Senna AS (2015) Isolation and characterization of a Siphoviridae phage infecting Bacillus megaterium from a heavily trafficked holy site in Saudi Arabia. Folia Microbiol 60:289–295. https://doi.org/10.1007/s12223-015-0375-1
Yang Y, Cai L, Ma R, Xu Y, Tong Y, Huang Y, Jiao NZ, Zhang R (2017) A novel Roseosiphophage isolated from the oligotrophic South China Sea. Viruses 9:1–16. https://doi.org/10.3390/v9050109
Kang I, Kang D, Cho JC (2016) Complete genome sequence of bacteriophage P2559Y, a marine phage that infects Croceibacter atlanticus HTCC2559. Marine Genom 29:35–38. https://doi.org/10.1016/j.margen.2016.07.001
Liu ZY, Wang M, Meng X, Li Y, Wang DB, Jiang Y, Shao HB, Zhang YY (2017) Isolation and genome sequencing of a novel Pseudoalteromonas phage PH1. Curr Microbiol 74:1–7. https://doi.org/10.1007/s00284-016-1175-9
Xu YQ, Ma YY, Yao S, Jiang ZY, Pei JS, Cheng C (2016) Characterization, genome sequence, and analysis of Escherichia phage CICC 80001, a bacteriophage infecting an efficient l-aspartic acid producing Escherichia coli. Food Environ Virol 8:18–26. https://doi.org/10.1007/s12560-015-9218-0
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P (2004) Structure of a bifunctional DNA primase-polymerase. Nat Struct Mol Biol 11:157–162. https://doi.org/10.1038/nsmb723
Gao Y, Liu Q, Wang M, Zhao GH, Jiang Y, Malin G, Gong Z, Meng X, Liu ZY, Lin TT, Li YT, Shao HB (2017) Characterization and genome sequence of marine Alteromonas gracilis phage PB15 isolated from the Yellow Sea, China. Curr Microbiol 74:821–826. https://doi.org/10.1007/s00284-017-1251-9
Yuan L, Cui Z, Wang Y, Guo X, Zhao Y (2014) Complete genome sequence of virulent bacteriophage SHOU24, which infects foodborne pathogenic Vibrio parahaemolyticus. Adv Virol 159:3089–3093. https://doi.org/10.1007/s00705-014-2160-x
Li P, Chen B, Song Z, Song Y, Yang Y, Ma P, Wang H, Ying J, Ren P, Yang L, Gao G, Jin S, Bao Q, Yang H (2012) Bioinformatic analysis of the Acinetobacter baumannii phage AB1 genome. Gene 507:125–134. https://doi.org/10.1016/j.gene.2012.07.029
Halgasova N, Mesarosova I, Bukovska G (2012) Identification of a bifunctional primase–polymerase domain of corynephage BFK20 replication protein gp43. Virus Res 163:454–460. https://doi.org/10.1016/j.virusres.2011.11.005
Black LW (1989) DNA Packaging in dsDNA bacteriophages. Annu Rev Microbiol 43:267–292. https://doi.org/10.1146/annurev.mi.43.100189.001411
Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G (2012) Small terminase couples viral DNA binding to genome-packaging atpase activity. Structure 20:1403–1413. https://doi.org/10.1016/j.str.2012.05.014
Li E, Xiao W, Ma Y, Zhe Y, Li H, Lin W, Wang X, Li C, Shen Z, Zhao R, Yang H, Jiang A, Yang W, Yuan J, Zhao X (2016) Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant enterobacter aerogenes. Sci Rep 6:28338. https://doi.org/10.1038/srep28338
Xu J, Hendrix RW, Duda RL (2004) Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell 16:11–21. https://doi.org/10.1016/j.molcel.2004.09.006
Kropinski AM, Waddell T, Meng J, Franklin K, Ackermann HW, Ahmed R, Mazzocco A, Yates J, Lingohr EJ, Johnson RP (2013) The host-range, genomics and proteomics of Escherichia coli, O157:H7 bacteriophage rV5. Virol J 10:1–12. https://doi.org/10.1186/1743-422X-10-76
Colangelo-Lillis JR, Deming JW (2013) Genomic analysis of cold-active Colwelliaphage 9A and psychrophilic phage-host interactions. Extremophiles 17:99–114. https://doi.org/10.1007/s00792-012-0497-1
Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM (2008) Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using blastp-based tools. Res Microbiol 159:406–414. https://doi.org/10.1016/j.resmic.2008.03.005
Acknowledgements
The research was funded by the Marine Scientific and Technological Innovation Project Financially supported by the Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0104-4, 2018SDKJ0406-6, and 2016ASKJ14), the National Natural Science Foundation of China (41676178, 31500339, and 41076088), the National Key Research and Development Program of China (2017YFA0603200 and 2018YFC1406704), the State Oceanic Administration People’s republic of China (GASI-02-PAC-ST-MSwin, GASI-02-PAC-ST-MSaut), and the Fundamental Research Funds for the Central University of Ocean University of China (201812002, 201762017, and 201562018).
Author information
Authors and Affiliations
Contributions
Hongbing Shao, Yong Jiang, Tong Jiang, Meiwen Wang, Yuye Han, and Min Wang conceived and designed the experiments and critically evaluated the manuscript. Xinran Zhang and Qi Wang conducted the sequencing experiments. Lei Zhao isolated and identified the phage and conducted the biological characterization experiments. Andrew McMinn helped to edit the language of manuscript. Yundan Liu was responsible for the data and sequence analyses and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Andrew Millard.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhao, L., Wang, M. et al. Complete genomic sequence of bacteriophage P23: a novel Vibrio phage isolated from the Yellow Sea, China. Virus Genes 55, 834–842 (2019). https://doi.org/10.1007/s11262-019-01699-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-019-01699-3