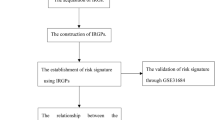
Abstract
Background
Bladder cancer, predominantly affecting men, is a prevalent malignancy of the urinary system. Although platinum-based chemotherapy has demonstrated certain enhancements in overall survival when compared to surgery alone, the efficacy of treatments is impeded by the unfavorable side effects of conventional chemotherapy medications. Nonetheless, immunotherapy exhibits potential in the treatment of bladder cancer.
Methods
To create an immune-associated prognostic signature for bladder cancer, bioinformatics analyses were performed utilizing The Cancer Genome Atlas (TCGA) database in this study. By identifying differential gene expressions between the high-risk and low-risk groups, a potential therapeutic drug was predicted using the Connectivity Map database. Subsequently, the impact of this drug on the growth of T24 cells was validated through MTT assay and 3D cell culture techniques.
Results
The signature included 1 immune-associated LncRNA (NR2F1-AS1) and 16 immune-associated mRNAs (DEFB133, RBP7, PDGFRA, CGB3, PDGFD, SCG2, ADCYAP1R1, OPRL1, PGR, PSMD1, TANK, PRDX1, ADIPOR2, S100A8, AHNAK, EGFR). Based on the assessment of risk scores, the patients were classified into cohorts of low-risk and high-risk individuals. The cohort with low risk demonstrated a considerably higher likelihood of survival in comparison to the group with high risk. Furthermore, variations in immune infiltration were noted among the two categories. Cephaeline, a possible medication, was discovered by analyzing variations in gene expression. It exhibited promise in suppressing the viability and growth of T24 bladder cancer cells.
Conclusion
The novel predictive pattern allows for efficient categorization of patients with bladder cancer, enabling focused and rigorous treatment for those expected to have a worse prognosis. The discovery of a possible curative medication establishes a basis for forthcoming immunotherapy trials in bladder cancer.













Similar content being viewed by others
Data Availability
The data of RNA-HTSeq and clinical information data of BLCA patients were derived from the following resources available in the public domain: https://portal.gdc.cancer.gov/; The data that support the findings of this study in Figure 13 are openly available in figshare at https://doi.org/10.6084/m9.figshare.24168177.
References
Black PC, Sridhar S (2017) New research in bladder cancer, ASCO-GU 2017. Can Urol Assoc J 11:S160–S162. https://doi.org/10.5489/cuaj.4606
Luo H, Xu C, Liu Z et al (2019) Neural differentiation of bone marrow mesenchymal stem cells with human brain-derived neurotrophic factor gene-modified in functionalized self-assembling peptide hydrogel in vitro. J Cell Biochem 120:2828–2835. https://doi.org/10.1002/jcb.26408
Arab S, Hadjati J (2019) Adenosine blockage in tumor microenvironment and improvement of cancer immunotherapy. Immune Netw 19:e23. https://doi.org/10.4110/in.2019.19.e23
Wolacewicz M, Hrynkiewicz R, Grywalska E et al (2020) Immunotherapy in bladder cancer: current methods and future perspectives. Cancers (Basel). https://doi.org/10.3390/cancers12051181
Londono MC, Reig M, Group RM (2020) Multidisciplinary clinical approach to cancer patients with immune-related adverse events induced by checkpoint inhibitors. Cancers (Basel). https://doi.org/10.3390/cancers12113446
Yang F, Zeng Z, Li J et al (2018) PD-1/PD-L1 axis, rather than high-mobility group alarmins or CD8+ tumor-infiltrating lymphocytes, is associated with survival in head and neck squamous cell carcinoma patients who received surgical resection. Front Oncol 8:604. https://doi.org/10.3389/fonc.2018.00604
Hu-Lieskovan S, Bhaumik S, Dhodapkar K et al (2020) SITC cancer immunotherapy resource document: a compass in the land of biomarker discovery. J Immunother Cancer. https://doi.org/10.1136/jitc-2020-000705
Linden A, Yarnold PR (2017) Modeling time-to-event (survival) data using classification tree analysis. J Eval Clin Pract 23:1299–1308. https://doi.org/10.1111/jep.12779
Nagashima K, Sato Y (2017) Information criteria for Firth’s penalized partial likelihood approach in Cox regression models. Stat Med 36:3422–3436. https://doi.org/10.1002/sim.7368
Tibshirani R (1997) The lasso method for variable selection in the Cox model. Stat Med 16:385–395. https://doi.org/10.1002/(sici)1097-0258(19970228)16:4%3c385::aid-sim380%3e3.0.co;2-3
Ritchie ME, Phipson B, Wu D et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25:25–29. https://doi.org/10.1038/75556
Kanehisa M, Furumichi M, Tanabe M et al (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucl Acids Res 45:D353–D361. https://doi.org/10.1093/nar/gkw1092
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Chen B, Khodadoust MS, Liu CL et al (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259. https://doi.org/10.1007/978-1-4939-7493-1_12
Musa A, Ghoraie LS, Zhang SD et al (2017) A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform 18:903. https://doi.org/10.1093/bib/bbx023
Han J, Gu X, Li Y et al (2020) Mechanisms of BCG in the treatment of bladder cancer-current understanding and the prospect. Biomed Pharmacother 129:110393. https://doi.org/10.1016/j.biopha.2020.110393
Pettenati C, Ingersoll MA (2018) Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat Rev Urol 15:615–625. https://doi.org/10.1038/s41585-018-0055-4
Gomes-Giacoia E, Miyake M, Goodison S et al (2014) Intravesical ALT-803 and BCG treatment reduces tumor burden in a carcinogen induced bladder cancer rat model; a role for cytokine production and NK cell expansion. PLoS ONE 9:e96705. https://doi.org/10.1371/journal.pone.0096705
Huntington ND, Legrand N, Alves NL et al (2009) IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J Exp Med 206:25–34. https://doi.org/10.1084/jem.20082013
Rosser CJ, Tikhonenkov S, Nix JW et al (2021) Safety, tolerability, and long-term clinical outcomes of an IL-15 analogue (N-803) admixed with bacillus calmette-guerin (BCG) for the treatment of bladder cancer. Oncoimmunology 10:1912885. https://doi.org/10.1080/2162402X.2021.1912885
Jacob JB, Jacob MK, Parajuli P (2021) Review of immune checkpoint inhibitors in immuno-oncology. Adv Pharmacol 91:111–139. https://doi.org/10.1016/bs.apha.2021.01.002
Alanee S, Sana S, El-Zawahry A et al (2021) Phase I trial of intravesical Bacillus Calmette-Guerin combined with intravenous pembrolizumab in recurrent or persistent high-grade non-muscle-invasive bladder cancer after previous Bacillus Calmette-Guerin treatment. World J Urol 39:3807–3813. https://doi.org/10.1007/s00345-021-03716-3
Ward Grados DF, Ahmadi H, Griffith TS et al (2022) Immunotherapy for bladder cancer: latest advances and ongoing clinical trials. Immunol Investig 51:2226–2251. https://doi.org/10.1080/08820139.2022.2118606
Inman BA, Longo TA, Ramalingam S et al (2017) Atezolizumab: a PD-L1-blocking antibody for bladder cancer. Clin Cancer Res 23:1886–1890. https://doi.org/10.1158/1078-0432.CCR-16-1417
Massard C, Gordon MS, Sharma S et al (2016) Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J Clin Oncol 34:3119–3125. https://doi.org/10.1200/JCO.2016.67.9761
Powles T, Park SH, Voog E et al (2020) Avelumab maintenance therapy for advanced or metastatic urothelial carcinoma. N Engl J Med 383:1218–1230. https://doi.org/10.1056/NEJMoa2002788
Feng L, Yang J, Zhang W et al (2022) Prognostic significance and identification of basement membrane-associated lncRNA in bladder cancer. Front Oncol 12:994703. https://doi.org/10.3389/fonc.2022.994703
Li DX, Feng DC, Shi X et al (2023) Identification of endothelial-related molecular subtypes for bladder cancer patients. Front Oncol 13:1101055. https://doi.org/10.3389/fonc.2023.1101055
Chen H, Yang W, Xue X et al (2022) Integrated analysis revealed an inflammatory cancer-associated fibroblast-based subtypes with promising implications in predicting the prognosis and immunotherapeutic response of bladder cancer patients. Int J Mol Sci. https://doi.org/10.3390/ijms232415970
Kashiwagi E, Abe T, Kinoshita F et al (2020) The role of adipocytokines and their receptors in bladder cancer: expression of adiponectin or leptin is an independent prognosticator. Am J Transl Res 12:3033–3045
Herik Rodrigo AG, Tomonobu N, Yoneda H et al (2022) Toll-like receptor 4 promotes bladder cancer progression upon S100A8/A9 binding, which requires TIRAP-mediated TPL2 activation. Biochem Biophys Res Commun 634:83–91. https://doi.org/10.1016/j.bbrc.2022.09.116
Yasar O, Akcay T, Obek C et al (2017) Significance of S100A8, S100A9 and calprotectin levels in bladder cancer. Scand J Clin Lab Investig 77:437–441. https://doi.org/10.1080/00365513.2017.1336567
Lyu ZJ, Wang Y, Huang JL et al (2021) Recurrent ZNF83-E293V mutation promotes bladder cancer progression through the NF-kappaB pathway via transcriptional dysregulation of S100A8. Mol Ther 29:275–290. https://doi.org/10.1016/j.ymthe.2020.09.004
Minami S, Sato Y, Matsumoto T et al (2010) Proteomic study of sera from patients with bladder cancer: usefulness of S100A8 and S100A9 proteins. Cancer Genom Proteom 7:181–189
Gao Z, Chen C, Gu P et al (2022) The tumor microenvironment and prognostic role of autophagy- and immune-related genes in bladder cancer. Cancer Biomark 35:293–303. https://doi.org/10.3233/CBM-220058
Luo Y, Chen L, Zhou Q et al (2020) Identification of a prognostic gene signature based on an immunogenomic landscape analysis of bladder cancer. J Cell Mol Med 24:13370–13382. https://doi.org/10.1111/jcmm.15960
Kriegmair MC, Wirtz RM, Worst TS et al (2018) Prognostic value of molecular breast cancer subtypes based on Her2, ESR1, PGR and Ki67 mRNA-expression in muscle invasive bladder cancer. Transl Oncol 11:467–476. https://doi.org/10.1016/j.tranon.2018.02.001
Salah Fararjeh A, Al-Khader A, Al-Saleem M et al (2021) The prognostic significance of proteasome 26S subunit, non-ATPase (PSMD) genes for bladder urothelial carcinoma patients. Cancer Inform 20:11769351211067692. https://doi.org/10.1177/11769351211067692
Kumari N, Vasudeva P, Tanwar P et al (2022) Peroxiredoxins—urinary surveillance biomarkers in urothelial cancer. J Cancer 13:2751–2756. https://doi.org/10.7150/jca.69811
Jin K, Qiu S, Jin D et al (2021) Development of prognostic signature based on immune-related genes in muscle-invasive bladder cancer: bioinformatics analysis of TCGA database. Aging Albany NY 13:1859–1871. https://doi.org/10.18632/aging.103787
Guo L, Wu Q, Ma Z et al (2021) Identification of immune-related genes that predict prognosis and risk of bladder cancer: bioinformatics analysis of TCGA database. Aging (Albany NY) 13:19352–19374. https://doi.org/10.18632/aging.203333
Silva LC, Borgato GB, Wagner VP et al (2022) Cephaeline is an inductor of histone H3 acetylation and inhibitor of mucoepidermoid carcinoma cancer stem cells. J Oral Pathol Med 51:553–562. https://doi.org/10.1111/jop.13252
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (82000230 to Gao Jinlai, 82104197 to Jin Jing), Science and Technology Bureau of Jiaxing (2020AY10021), Basic Public Welfare Research Project of Zhejiang Province (LGC21B050011) and the Qin Shen Scholar Program of Jiaxing University. We thank Jiaxing Medical Key Subject Funding of Zhejiang Province (2023-ZC-013), the Jiaxing Key Laboratory of Precise Diagnosis and Treatment of Urological Tumor (2020-mnzdsys) and Jiaxing Innovation Leading Program for Elite Talents (84122009) for their help.
Author information
Authors and Affiliations
Contributions
The study was conceptualized by ZX and LZ. The experiments and data analysis were conducted by PH, SB, FX, WJ, ZB, and LS. The original draft of the paper was written by JJ, HY, and GJ. Constructive discussions involving JJ and GJ contributed to the enhancement of the paper. The final manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Conflict of interest
No competing interests are declared by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11255_2023_3796_MOESM1_ESM.pdf
Figure S1. Clinical information varied between the high and low-risk groups in the risk test database. Figure S2. Clinical information varied between the high and low-risk groups in the risk train database. Figure S3. Heatmap of immune cell expression between high-risk and low-risk groups. Figure S4. Barplot of immune cell expression between high-risk and low-risk groups (PDF 280 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiaoqin, Z., Zhouqi, L., Huan, P. et al. Development of a prognostic signature for immune-associated genes in bladder cancer and exploring potential drug findings. Int Urol Nephrol 56, 483–497 (2024). https://doi.org/10.1007/s11255-023-03796-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11255-023-03796-7