Abstract
Phosphoethanolamine N-methyltransferase (PEAMTase) catalyzes the methylation of phosphoethanolamine to produce phosphocholine and plays an important role in the abiotic stress response. Although the PEAMT genes has been isolated from many species other than pitaya, its role in the drought stress response has not yet been fully elucidated. In the present study, we isolated a 1485 bp cDNA fragment of HpPEAMT from pitaya (Hylocereus polyrhizus). Phylogenetic analysis showed that, during its evolution, HpPEAMT has shown a high degree of amino acid sequence similarity with the orthologous genes in Chenopodiaceae species. To further investigate the function of HpPEAMT, we generated transgenic tobacco plants overexpressing HpPEAMT, and the transgenic plants accumulated significantly more glycine betaine (GB) than did the wild type (WT). Drought tolerance trials indicated that, compared with those of the wild-type (WT) plants, the roots of the transgenic plants showed higher drought tolerance ability and exhibited improved drought tolerance. Further analysis revealed that overexpression of HpPEAM in Nicotiana tabacum resulted in upregulation of transcript levels of GB biosynthesis-related genes (NiBADH, NiCMO and NiSDC) in the leaves. Furthermore, compared with the wild-type plants, the transgenic tobacco plants displayed a significantly lower malondialdehyde (MDA) accumulation and higher activities of the superoxide dismutase (SOD) and peroxidase (POD) antioxidant enzymes under drought stress. Taken together, our results suggested that HpPEAMT enhanced the drought tolerance of transgenic tobacco.
Key message
Overexpression of the pitaya phosphoethanolamine N-methyltransferase gene (HpPEAMT) enhanced simulated drought stress in tobacco.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought stress is one of the most important abiotic stresses that affects crop yields throughout the world (Thirunavukkarasu et al. 2017). One strategy for improving crop tolerance to drought stress is to introduce or increase the capability to synthesize osmoprotectants such as soluble sugars, proline and inositol (Ahammed et al. 2020a). Glycine betaine (GB) is recognized as the most effective osmoprotectant that confers tolerance to drought stresses (Zhang et al. 2011; Nawaz and Wang 2020). Moreover, it can stabilize protein and membrane structures (Gao et al. 2019). Several studies have reported the existence of a positive correlation between exogenous GB applications and tolerance to drought stress in crops (Annunziata et al. 2019; Tisarum et al. 2019). GB is widely present in cyanobacteria, algae, bacteria, animals, and higher plants (Chen and Murata 2011; Scholz et al. 2016). Some plant species of the Chenopodiaceae family, such as spinach, barley, and beets, can maintain the osmotic pressure of cells by synthesizing and accumulating a large amount of GB to alleviate the damage of drought stresses, and some plants of the Poaceae family, such as maize, wheat, sorghum, and barley can synthesize and accumulate detectable GB (Yu et al. 2014). These studies strongly demonstrated that GB plays an important role in tolerance to drought stress.
GB is synthesized from choline catalyzed by choline monooxygenase (CMO) and then catalyzed by betaine aldehyde hydrogenase (BADH) (Razavi et al. 2018). In plants, previous studies have shown that deficiency of choline, the substrate of GB biosynthesis, limits the accumulation of GB and PEAMTase is the limiting key enzyme involved in choline synthesis (McNeil et al. 2000). Firstly, serine is decarboxylated by serine decarboxylase (SDC) to produce ethanolamine, and then three methylation steps from phosphoethanolamine, a derivative of ethanolamine to phosphocholine are all catalyzed by PEAMTase (Fig. S1). The phosphocholine produce choline and phosphatidic acid under the hydrolysis of phosphocholine phosphatase (Li et al. 2016; Niu et al. 2018a). The PEAMT gene was first isolated from Arabidopsis (Bolognese and Mcgraw 2000). The PEAMT has subsequently been cloned from spinach (Nuccio et al. 2000), Triticum aestivum (Charron et al. 2002), Atriplex nummularia (Tabuchi et al. 2005), Beta vulgaris (Tabuchi et al. 2006), Zea mays (Wu et al. 2007) and green algae (Hirashima et al. 2017). To date, there has been no report concerning PEAMT gene isolation from pitaya.
Pitaya (Hylocereus polyrhizus), also known as dragon fruit, belongs to the Hylocereus genus, which is a member of the Cactaceae family (Ortiz and Takahashi 2015). The growing area of pitaya is expanding rapidly in many tropical and subtropical areas worldwide, not only because of the exotic appearance and striking colors of the fruits but also because of their nutritional value and health properties (Hua et al. 2018; Ren et al. 2018). Moreover, pitaya demonstrates a high tolerance to drought stress (Nie et al. 2019) and is an excellent species for mining plant drought tolerance genes. In our laboratory, we preliminarily screened the PEAMT gene from pitaya by suppression subtractive hybridization and cDNA microarray approaches, confirming its response to drought stress (Fan et al. 2014). Previous studies on the function of PEAMT gene mostly focused on salt tolerance (Li et al. 2016; Wu et al. 2007). However, plant with strong salt tolerance does not mean it has a strong drought tolerance (such as salt-tolerant rice cultivar). Salt tolerance mechanism in plants is different from that of drought tolerance (Liang et al. 2018). Whether overexpression of pitaya PEAMT gene can cause the increase of GB content, whether it can improve the drought resistance of crops and the possible reasons for the improvement of drought resistance have yet been unknown so far. So for the first time, we cloned the pitaya PEAMT gene and examined the function of this gene by overexpressing it in tobacco. The aims of the present work were therefore (1) to verify the drought tolerance ability of transgenic lines overexpressing HpPEAMT, (2) to examine the role of HpPEAMT in GB accumulation in transgenic lines, and (3) to reveal the effect of HpPEAMT on the transcript levels of GB biosynthesis related genes. The results may be beneficial for further determining the role of HpPEAMT in the drought tolerance response as well as for determining a new strategy aimed at the engineering of plants with greater capability against abiotic stress.
Materials and methods
Plant materials and growth conditions
Pitaya (H. polyrhizus ‘Zihonglong’) (Nie et al. 2019) and tobacco (Nicotiana tabacum ‘Xanthinc’) plants were obtained and then conserved in the Key Laboratory of Plant Resources Conservation and Germplasm Innovation in the Mountainous Region (Ministry of Education), Institute of Agro-Bioengineering. All the plants were grown in an artificial climate chamber at 25/25 °C (day/night), under a 12/12 h (day/night) photoperiod provided by supplementary lighting with a 300 μmol/(m2 s) light intensity and under 60% relative humidity.
Cloning and bioinformatic analysis of HpPEAMT
Total RNA was isolated from fleshy stems of pitaya using a TransZol Plant RNA Extraction Kit (Tiangen, Beijing, China), and cDNA was synthesized using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China) according to the manufacturer’s instructions. The cDNA was then diluted ten times with ddH2O. HpPEAMT was obtained from the diluted cDNA by using a pair of primers (HpPEAMT-F and HpPEAMT-R) based on pitaya transcriptomic data in our lab and on a published sequence (accession No. XP_021715259.1). The sequences of the primers used were as follows: HpPEAMT-F (5′-ATGGCTGCTCAAGAAATGGCAATGGC-3′) and HpPEAMT-R (5′-TCAGTGTTTCTTGGCAATGAACAAAC-3′). PCR amplification was performed with PrimeSTAR Max DNA Polymerase (TaKaRa, Dalian, China), with initial denaturation at 94 °C for 4 min followed by 35 cycles of 94 °C for 30 s, 54 °C for 30 s and 72 °C for 90 s. Afterward, there was a final extension at 72 °C for 10 min. The PCR products were cloned into a pEASY®-Blunt Simple Cloning vector and then transformed into Escherichia coli DH5α cells. The resulting transformants were selected on agar plates [Luria Broth (LB), 1.5% (w/v) agar, Kan (50 μg/mL)]. The isolated sequences were subsequently sequenced by Sangon Biotech Corporation (Shanghai, China).
The deduced amino acid sequence of HpPEAMT was queried against the NCBI Conserved Domain Database to identify the conserved methyltransferase domains. PEAMT protein sequences from other plant species were obtained from the GenBank database. All the sequences were aligned by DNAMAN software before evolutionary analysis with Molecular Evolutionary Genetics Analysis (MEGA) 5.2 software, during which the neighbor-joining (NJ) method was used to infer and construct a phylogenetic tree (Kumar et al. 2008).
Generation of HpPEAMT transgenic tobacco plants
The cDNA sequence of HpPEAMT was amplified using a pair of primers (HpPEAMT-T-F and HpPEAMT-T-R) harboring the Bam HI and Sac I restriction sites. The sequences of these primers were as follows: HpPEAMT-T-F (5′-TTTACAATTACCATGG/GATCCATGGCTGCTCAAGAAATGGCAATGGC-3′, Bam HI site in italics) and HpPEAMT-T-R (5′- ACCGATGATACGAACGAGCT/CTCAGTGTTTCTTGGCAATGAACAAAC-3′, Sac I site in italics). The amplified product was purified and inserted into a pYN7736 vector containing the CaMV 35S promoter and GUS gene using a ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer’s instructions. The resultant construct was named pYN7736-HpPEAMT. Subsequently, the construct was verified by DNA sequencing before being transferred into Agrobacterium tumefaciens strain GV3101 with the freeze–thaw method. The GV3101 strain containing the pYN7736-HpPEAMT construct was then used to transform tobacco (N. tabacum ‘Xanthinc’) by the leaf disc method (Horsch et al, 1985). Agar-solidified MS (Murashige and Skoog 1962) medium containing 25 mg/L hygromycin was used to screen T0, T1 and T2 transgenic lines. The transgenic status of T2 transgenic lines was verified based on GUS staining (Zhang et al. 2008), PCR amplification with a pair of primers: 7736-F (vector sequence, 5′-TTTACAATTACCATGGGATCC-3′) and HpPEAMT-R (HpPEAMT sequence, 5′-TCAGTGTTTCTTGGCAATGAACAAAC-3′) and HpPEAMT relative expression analysis.
Measurement of GB content
The GB content in the leaves of three T2 transgenic lines and WT were measured with high-performance liquid chromatography (HPLC) as previously described, with slight modifications (Luo et al. 2012). In brief, 100 mg of fresh leaves was cleaned, ground to a fine powder, extracted with 1 mL methanol overnight, and then centrifuged to collect the supernatant. The supernatant was subsequently dried under nitrogen, dissolved in the mobile phase to a constant volume (0.5 mL), and ultimately filtered through a 0.22 µm PTFE membrane before being injected into the HPLC instrument.
The GB content was quantified using an Agilent 1100 series (Agilent Technologies, Santa Clara, CA, USA) automated HPLC machine. The HPLC system consisted of a Kromasil C18 reversed-phase column (250 mm × 4.6 mm, 5 µm), a potassium dihydrogen phosphate:water (0.68:100 m/v; pH 4–5, with phosphoric acid) mixture as the mobile phase, a detection wavelength of 195 nm, 0.5 mL/min flow rate, a column temperature of 30 °C and an injection volume of 10 µL. All the data were quantified on the basis of their standard curves.
Drought tolerance assays
Three T2 transgenic lines (L10, L11, L16) and WT were used for drought stress treatments. First, the seeds were sterilized and then sown into MS media with different mannitol concentrations (150 and 300 mmol/L) for analysis of root drought tolerance. MS media lacking mannitol served as the control. Nine plants from each line were selected to measure the root length after 21 days. After seeds normally growing for 14 days in nutrient-enriched soil, a total of 18 plants (nine for WT plants and nine for transgenic plants) of similar height and health conditions were irrigated with PEG 6000 (− 0.60 MPa) to simulate drought stress, which was conducted once every other day for 8 days. The seedlings not treated with PEG were used as the control (− 0.01 MPa). The Water Potential was determined by a WP4C PotentiaMeter (METER Group Inc. Pullman, WA, USA). The third or fourth leaf from the bottom to the top was harvested from each plant at 0, 4, and 8 days under drought stress, immediately frozen in liquid nitrogen and then stored at − 80 °C for qRT-PCR, MDA content and antioxidant activities quantifications. Total chlorophyll contents of each plant were measured by the method described by You et al (2019). All experiment was repeated at least three times. The growth status of the plants was also recorded with images.
Quantification of MDA content and antioxidant activities
The MDA content and antioxidant activities, including those of SOD and POD, of each plant were measured by commercial assay kits (Suzhou Keming Biotechnology Co., Ltd., China) according to the manufacturer's instructions (Bai et al. 2018).
Expression profiles of GB biosynthesis-related genes
Expression profiles of HpPEAMT and GB biosynthesis-related genes (Fig. S1) in the WT and T2 transgenic lines were measured through quantitative real-time polymerase chain reaction (qRT-PCR). qRT-PCR was carried out using an ABI 7500 real-time PCR system (Applied Biosystems, USA) and a real-time PCR kit SYBR Premix Ex Taq Kit (TaKaRa). The β-Actin gene (accession No. AB158612) was used as an internal control for normalization. Table S1 shows the primer sequences used for qRT-PCR.
Data analysis
All the data were expressed as the means ± SEs. Significant differences were assessed by one-way ANOVA followed by post hoc Duncan’s multiple range tests (p < 0.05 or p < 0.01). All analyses were performed with DPS 7.05 software (China) and Microsoft Excel 2007.
Results
Identification and bioinformatic analysis of HpPEAMT
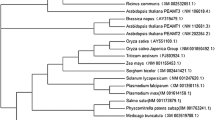
In pitaya, the open reading frame (ORF) length of HpPEAMT was 1485 bp (Table S2) and encodes 494 putative amino acids (Fig. 1). The deduced HpPEAMT amino acid sequences contained two tandem methyltransferase domains, which were located at the N-terminus (60–158 aa) and the C-terminus (289–386 aa). Each domain contained four motifs labeled I, post-I, II, or III (Fig. 1). The deduced HpPEAMT amino acid sequence shared 84.62% identity with the published amino acid sequence (XP_021715259.1) (Table S3). To compare the molecular evolutionary relationships of HpPEAMT with PEAMT genes of other species, we downloaded 30 sequences encoding HpPEAMT from the GenBank database (Table S3), and the phylogenetic analysis revealed that the HpPEAMT protein sequences in pitaya and those of the Chenopodiaceae species (Atriplex canescens, Spinacia oleracea, Chenopodium quinoa, Beta vulgaris, Salicornia europaea, and Suaeda liaotungensis) clustered together, forming a monophyletic branch (Fig. 2).
Alignment of deduced HpPEAMT amino acid sequences with those of the PEAMT ortholog from Chenopodium quinoa (accession No. XP_021715259.1). Sequence motifs defining the methyltransferase domains are labeled I, post-I, II, and III. The motifs in the N-terminal and C-terminal methyltransferase domains are denoted with red and blue boxes, respectively. (Color figure online)
Evolutionary analysis of HpPEAMT amino acid sequences. The HpPEAMT and PEAMT protein sequences from other plant species were aligned before evolutionary analysis. Evolutionary analysis was performed with molecular evolutionary genetics analysis (MEGA) 5.2 software, using the Neighbor-Joining (NJ) method to infer and construct phylogenetic tree. Bootstrap test: 1000 bootstrap replicates; Poisson model
Generation of transgenic tobacco plants
T0 positive plants were self-pollinated to produce T1 seeds. T1 positive plants were self-pollinated to produce T2 seeds.
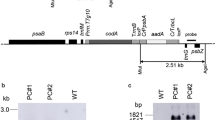
Compared with WT, after screening with 25 mg/L hygromycin for 16 days. The positive rates of three T2 generation lines (L10, L11 and L16) were 100% and could be regarded as homozygous lines (Fig. S2). Three T2 transgenic lines (L10, L11, L16) were then identified via GUS staining, PCR and HpPEAMT relative expression. GUS activity (Fig. 3a) and the expected 1485 bp amplified fragment (Fig. 3b), which was not detected in the WT, were detected in the leaves of the transgenic lines. The relative expression of HpPEAMT in the transgenic lines was significantly higher (p < 0.01) than that of the WT (Fig. 3c), indicating that HpPEAMT had been integrated into the tobacco genome [compared with that of HpPEAMT, the relative expression of the tobacco homologous gene (XM_016581921.1) was very low in both the transgenic and WT lines and was negligible].
Identification of T2 transgenic lines (L10, L11 and L16) through a GUS staining, b PCR and c HpPEAMT overexpression levels. Data are the means ± SEs of three biological replicates; asterisks indicate significant differences (*p < 0.05, **p < 0.01) in comparison with the wild type. The scale bar represents 0.5 cm
HpPEAMT increased GB content in transgenic tobacco
To determine whether the insertion of HpPEAMT into the tobacco genome resulted in the enhancement of GB accumulation, the GB contents of three T2 transgenic lines were analyzed quantitatively by HPLC, a highly sensitive detection method. The amounts of GB accumulated in the leaves of T2 transgenic lines were 0.29, 0.48, and 0.24 nmol/g FW (fresh weight), respectively, which were 2.07, 3.43 and 1.71 times the amount in the WT (0.14 nmol/g FW) (Fig. 4).
HpPEAMT enhances drought tolerance of transgenic tobacco
Both WT and transgenic seedlings grew in vertical petri dishes with different concentrations of mannitol for 21 days, and the root growth was overall inhibited to some extent with increasing mannitol concentration. However, the root growth of WT was more significantly (p < 0.05) inhibited than that of the transgenic lines. Specifically, there was no significant difference between the WT and transgenic lines in the absence of mannitol (Fig. 5a, a′). Under treatment with 150 mmol/L mannitol, the root lengths of the three transgenic lines (L10, L11 and L16) were 38, 24, and 12% longer, respectively, than those of the WT (Fig. 5b, b′). Following treatment with 300 mmol/L mannitol, the root lengths of the three transgenic lines were 47, 37, and 44% significantly longer, respectively, than those of the WT plants (Fig. 5c, c′). The results indicated that the transgenic lines had a more developed root system than the WT under drought stress. Morphologically, there was no significant difference between the WT and transgenic lines under nonstressed conditions (Fig. 5d). However, the WT plants were slightly wilted and yellow after 4 days of drought stress (Fig. 5e) and were extensively wilted and yellow when treated for 8 days of drought stress (Fig. 5f), while the transgenic lines were hardly injured and still healthy. There was no significant difference in the chlorophyll content between WT and transgenic lines under nonstressed conditions (Fig. 5d′). But under drought stress, the chlorophyll content of transgenic lines was higher than that of WT plants while the chlorophyll content of WT plants decreased significantly (Fig. 5e′, f′). Together, these results suggested that the transgenic plants exhibited stronger resistance to drought stress.
Phenotypic differences between the WT and transgenic tobacco plants under drought stress. a–c′ Root growth status of WT and T2 transgenic lines (L10, L11 and L16) exposed to different mannitol concentrations. The scale bar represents 1 cm; d–f and d′–f′ are respectively growth status and chlorophyll content of WT and T2 transgenic lines (L10, L11 and L16) exposed to PEG (− 4.9 MPa) at different time points. Data are the means ± SEs of three biological replicates; asterisks indicate significant differences (*p < 0.05, **p < 0.01) in comparison with the wild type. The scale bar represents 10 cm
HpPEAMT alters MDA accumulation and antioxidant activities
To further investigate the role of HpPEAMT in antioxidant defense under nonstressed and drought-stress conditions, the MDA content and antioxidant enzyme activities, including those of SOD and POD, in three T2 transgenic lines and WT were analyzed. There were no significant differences in MDA content between the WT and transgenic lines under nonstressed conditions. However, under 4 and 8 days of drought stress (Fig. 6a), the MDA content of the WT plants was significantly higher than that of transgenic lines (p < 0.01). Under nonstressed conditions, the activities of SOD in transgenic line L10 and L11 were 30 and 26% significantly higher than that of WT plants, respectively (Fig. 6b), and the activity of POD in transgenic line L10 was 21% significantly higher than that of WT plants (Fig. 6c). Under drought stress conditions (Fig. 6b, c), transgenic lines had significantly higher activities of both SOD and POD enzymes than did the WT (p < 0.01) except POD activity in transgenic line L16 under 4 days of drought stress. Together, these results showed that overexpression of HpPEAMT in tobacco could delay the accumulation of MDA in lipid membrane oxidation products and significantly increase antioxidant enzyme activity.
MDA (a) content and SOD (b) and POD (c) activity in WT and T2 transgenic lines (L10, L11 and L16) exposed to PEG (− 4.9 MPa) at different time points. Data are the means ± SEs of three biological replicates; asterisks indicate significant differences (*p < 0.05, **p < 0.01) in comparison with the wild type
HpPEAMT influences the transcript levels of GB biosynthesis related genes
We analyzed the expression levels of GB biosynthesis-related genes in the leaves of the T2 transgenic lines and WT by qRT-PCR experiments. The relative expression of the NiBADH in the transgenic lines was significantly higher that of WT under both nonstressed and drought-stress conditions (Fig. 7a). Compared with that of WT plants, the relative expression of NiCMO in transgenic lines was significantly higher under drought-stress conditions, and oppositly, the relative expression of the NiCMO in transgenic lines was significantly lower under nonstressed conditions (Fig. 7b). The relative expression of the NiSDC was not significantly different between the WT and transgenic lines under nonstressed conditions (Fig. 7c). However, after treatment for 4 days, the levels in transgenic lines L11 and L16 peaked at values approximately 4.37 and 2.58 times those of the WT (p < 0.01) and then decreased at 8 days (Fig. 7c). Together, these results indicated that overexpression of HpPEAMT in tobacco could significantly upregulate the expression levels of GB biosynthesis-related genes under drought stress conditions.
Expression quantification (via qRT-PCR) of GB biosynthesis-related genes in WT and T2 transgenic lines (L10, L11 and L16) subjected to PEG (− 4.9 MPa) at different time points. a NiBADH, b NiCMO and c NiSDC. Data are the means ± SEs of three biological replicates; asterisks indicate significant differences (*p < 0.05, **p < 0.01) in comparison with the wild type
Discussion
As a member of the family Cactaceae, pitaya is highly tolerant to drought stress and is recognized as a promising crop species for drought-stricken areas (Nie et al. 2019). Available evidence has indicated that the HpPEAMT is significantly upregulated in pitaya under drought stress (Fan et al. 2014), and the same phenomenon has also been documented in red seaweed (Mao et al. 2019). Therefore, PEAMT is possibly responsive to drought stress and may be useful in genetic engineering to improve the drought tolerance ability of plants. To assess whether HpPEAMT enhanced the drought tolerance of transgenic tobacco plants, three T2 transgenic lines (L 10, L 11 and L 16) with high expression levels for root length measurements and quantification of antioxidant activities and MDA content. Root system is the first part of the plant that responds to drought and simultaneously sends a signal to the aboveground parts to induce a physiological response to improve the ability of plants to resist drought. Therefore, a well-developed root system can effectively increase the rational distribution of water to plants, reduce the damage caused by drought, and increase the survival rate under stress conditions (Xu et al. 2018). The results of the root drought resistance-ability test showed that the root growth of the three transgenic lines was less inhibited than was that of the WT under the treatment with high mannitol concentrations (Fig. 5a–c′), suggesting that the HpPEAMT transgenic plants exhibited stronger resistance to drought stress.
Because of their sessile lifestyle, plants have developed various strategies to survive and alleviate the negative impacts of abiotic stresses (Ahammed et al. 2020b; Ganie and Ahammed 2021). Among these strategies, the enzymatic antioxidant defense system plays an important role in protecting plant cells against oxidative damage by scavenging excess ROS, which largely accumulate in plants under abiotic stress conditions (Ahammed et al. 2020a, c). It has been demonstrated that drought tolerance is positively correlated with the increasing activities of antioxidant enzymes such as SOD, CAT and POD in plants (Li et al. 2019; Niu et al. 2018b). Furthermore, studies have reported that, compared with WT plants, transgenic plants with higher antioxidant enzyme activity had higher tolerance to drought stress (El-Esawi and Alayafi 2019), which is consistent with our current research results. Our study showed that the antioxidant enzyme activity of the HpPEAMT transgenic lines was significantly enhanced compared with that of the WT under both nonstressed and drought-stress conditions (Fig. 6b, c). MDA content is the final product of membrane lipid peroxidation in plant cell and often causes an increase in plant leaf electrical conductivity, so MDA content is widely considered an important physiological marker of membrane system injury under adverse stresses (Golldack et al. 2014; Xu et al. 2018). Guo et al. (2017) found that MDA content was negatively correlated with plant drought resistance. Our study showed that the MDA content in WT plants was significantly higher than that in the transgenic lines under drought-stress conditions (Fig. 6a). Overexpression of HpPEAMT increased the drought tolerance of the transgenic plants by decreasing the production of MDA and increasing the activities of antioxidant enzymes.
In addition to increasing their antioxidant enzyme activity to scavenge reactive oxygen species (Zhang et al. 2016), plants have also evolved protective strategies to cope with adverse environmental conditions by altering the expression of stress response genes controlling the production of proteins and metabolites. GB biosynthesis-related genes play an important role in improving the abiotic stress tolerance of transgenic plants (Chen and Murata 2011; Wei et al. 2017). Overall, our results showed that NiBADH, NiCMO and NiSDC, which are GB biosynthesis related genes in tobacco, were significantly upregulated in the HpPEAMT transgenic lines compared with the WT under drought stress (Fig. 7a–c). Thus, HpPEAMT overexpression and upregulated expression of GB biosynthesis related genes contributed to the increased GB content in transgenic plants. Not all genes involved in the GB biosynthesis pathway can increase the GB content of transgenic tobacco. The GB accumulation and stress resistance of transgenic tobacco overexpressing both CMO and BADH genes were lower than expected (McNeil et al. 2001). Endogenous choline levels are a limiting factor for tobacco that do not accumulate GB (Nuccio et al. 1998). Via an engineered GB pathway, overexpressing SpPEAMT in transgenic tobacco increased the levels of choline. Moreover, an expanded choline pool led to a further increase in the synthesis of GB (McNeil et al. 2001). In this study, when HpPEAMT was overexpressed in tobacco, the content of GB significantly increased compared to that in the WT (Fig. 4), which once again showed the importance of PEAMT in the synthesis of GB.
It has been well documented that even slight GB accumulation in some transgenic plants is enough to improve abiotic stress tolerance. For example, a slight GB accumulation (35 ± 15 nmol/g FW) improved salt tolerance of transgenic tobacco expressing betA, a gene from E. coli (Holmström et al. 2000). Similarly, less than 0.5 nmol/g FW GB content was apparently sufficient to increase tolerance to salt stress in tobacco expressing OsCMO, a gene from rice (Luo et al. 2012). On the other hand, Physiological studies have suggested that GB acts as an osmolyte for osmotic adjustment to maintain cellular water balance only when GB accumulates at high levels, e.g., 40 µmol/g FW under stress conditions (Rhodes and Hanson 1993; Al Hassan et al. 2016). In fact, The osmoregulation of osmolyte is related to the water potential gradient between the cell and the surrounding environment, as water always flows from high water potential to low water potential (Verslues et al. 2006). The highest GB content in our study in the transgenic plants was 0.48 nmol/g fresh weight. The difference in GB content in plants was probably due to the different GB measurement and calculation methods used (You et al. 2019) or may also be related to the different tobacco varieties, growth conditions and sampling locations. Many studies have shown that exogenous GB applications specifically improved the activity of antioxidant enzymes such as SOD and CAT in Lolium perenne (Hu et al. 2012), POD in Vigna unguiculata (Raza et al. 2007) and reduced MDA content in Glycine max (Malekzadeh 2015). Our results showed that, compared with WT plants, the transgenic tobacco plants exhibited higher GB content (Fig. 4), antioxidant enzyme activities (Fig. 6b, c), and drought tolerance (Fig. 5a–f′). Whether endogenous GB improved the antioxidant enzyme system of transgenic plants and thus enhanced their drought resistance and the mechanism behind the drought tolerance in the transgenic lines might need to be further studied by carrying out a series of technologies. In addition, although the transgenic tobacco plants overexpressing HpPEAMT showed better drought tolerance, pitaya and tobacco are organisms of different origins. The efficacy of GB engineering for important cultivated field crop species has never been demonstrated (Annunziata et al. 2019). In the future, additional work must be performed to collect data on the molecular mechanisms underlying the HpPEAMT response to drought stress and to determine whether HpPEAMT obtained from pitaya can also be applied to other crop species.
Conclusions
In summary, a 1485 bp cDNA fragment of HpPEAMT was isolated from pitaya (H. polyrhizus ‘Zihonglong’). Phylogenetic analysis showed that HpPEAMT had a high degree of amino acid sequence similarity with orthologous genes in Chenopodiaceae species throughout evolution. The drought tolerance trial indicated that the drought-tolerance ability of the roots of the transgenic lines was obviously higher than that of the WT at the seedling stage. Moreover, overexpression of HpPEAMT in the transgenic lines upregulated the transcript levels of GB biosynthesis related genes, led to an increased GB content and improved antioxidant enzyme activities, thus conferring drought stress tolerance to the transgenic lines.
Abbreviations
- APX:
-
Ascorbate peroxidase
- BADH :
-
Betaine aldehyde dehydrogenase gene
- CAT:
-
Catalase
- CMO :
-
Choline monooxygenase gene
- GB:
-
Glycine betaine
- GUS:
-
β-Glucuronidase
- MDA:
-
Malondialdehyde
- PEAMTase:
-
Phosphoethanolamine N-methyltransferase
- PEG:
-
Polyethylene glycol
- POD:
-
Peroxidase
- ROS:
-
Reactive oxygen species
- SDC :
-
Serine decarboxylase gene
- SOD:
-
Superoxide dismutase
- WT:
-
Wild type
References
Ahammed GJ, Li X, Wan HJ, Zhou GZ, Cheng Y (2020a) SlWRKY81 reduces drought tolerance by attenuating proline biosynthesis in tomato. Sci Hortic 2020:270. https://doi.org/10.1016/j.scienta.2020.109444
Ahammed GJ, Li X, Liu AR, Chen SC (2020b) Brassinosteroids in plant tolerance to abiotic stress. J Plant Growth Regul 3:39. https://doi.org/10.1007/s00344-020-10098-0
Ahammed GJ, Li C, Li X, Liu A, Chen S, Zhou J (2020c) Overexpression of tomato RING E3 ubiquitin ligase gene SlRING1 confers cadmium tolerance by attenuating cadmium accumulation and oxidative stress. Physiol Plantarum. https://doi.org/10.1111/ppl.13294
Al Hassan M, Chaura J, López-Gresa MP, Borsai O, Daniso E, Donat-Torres MP, Mayoral O, Vicente O, Boscaiu M (2016) Nativeinvasive plants vs. halophytes in Mediterranean salt marshes: stress tolerance mechanisms in two related species. Front Plant Sci 7:473. https://doi.org/10.3389/fpls.2016.00473
Annunziata MG, Ciarmiello LF, Woodrow P, Dell’Aversana E, Carillo P (2019) Spatial and temporal profile of glycine betaine accumulation in plants under abiotic stresses. Front Plant Sci 10:230. https://doi.org/10.3389/fpls.2019.00230
Bai L, Liu Y, Mu Y, Anwar A, He C, Yan Y, Li Y, Yu X (2018) Heterotrimeric G-protein γ subunit CsGG3.2 positively regulates the expression of CBF genes and chilling tolerance in cucumber. Front Plant Sci 9:488. https://doi.org/10.3389/fpls.2018.00488
Bolognese CP, Mcgraw P (2000) The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine: phospho-ethanolamine N-methyltransferase. Plant Physiol 124(4):1800–2181. https://doi.org/10.1104/pp.124.4.1800
Charron JB, Breton G, Danyluk J, Muzac I, Ibrahim RK, Sarhan F (2002) Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol 129(1):363–373. https://doi.org/10.1104/pp.001776
Chen TH, Murata N (2011) Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ 34(1):1–20. https://doi.org/10.1111/j.1365-3040.2010.02232.x
El-Esawi MA, Alayafi AA (2019) Overexpression of StDREB2 transcription factor enhances drought stress tolerance in cotton (Gossypium barbadense L.). Genes 10(2):142. https://doi.org/10.3390/genes10020142
Fan QJ, Yan FX, Qiao G, Zhang BX, Wen XP (2014) Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene 533(1):322–331. https://doi.org/10.1016/j.gene.2013.08.098
Gao Y, Li M, Zhang X, Yang Q, Huang B (2019) Up-regulation of lipid metabolism and glycine betaine synthesis are associated with choline-induced salt tolerance in halophytic seashore paspalum. Plant Cell Environ 43(1):159–173. https://doi.org/10.1111/pce.13657
Ganie SA, Ahammed GJ (2021) Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. https://doi.org/10.1007/s00299-020-02649-2
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151. https://doi.org/10.3389/fpls.2014.00151
Guo XY, Zhang L, Zhu JB, Wang AY, Liu HL (2017) Christolea crassifolia HARDY gene enhances drought stress tolerance in transgenic tomato plants. Plant Cell Tissue Org 129(3):469–481. https://doi.org/10.1007/s11240-017-1192-9
Hirashima T, Toyoshima M, Moriyama T, Nakamura Y, Sato N (2017) Characterization of phosphoethanolamine-N-methyltransferases in green algae. Biochem Biophys Res Commun 488(1):141–146. https://doi.org/10.1016/j.bbrc.2017.05.026
Holmström KO, Somersalo S, Mandal A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot 51(343):177–185. https://doi.org/10.1093/jexbot/51.343.177
Horsch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227(4691):1229–1231. https://doi.org/10.1126/science.227.4691.1229
Hu LX, Hu T, Zhang XZ, Pang HC, Fu JM (2012) Exogenous glycine betaine ameliorates the adverse effect of salt stress on perennial ryegrass. J Am Soc Hortic Sci 137(1):38–46. https://doi.org/10.21273/JASHS.137.1.38
Hua Q, Chen C, Tel Zur N et al (2018) Metabolomic characterization of pitaya fruit from three red-skinned cultivars with different pulp colors. Plant Physiol Biochem 126:117–125. https://doi.org/10.1016/j.plaphy.2018.02.027
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306. https://doi.org/10.1093/bib/bbn017
Liang W, Ma X, Wan P, Liu L (2018) Plant salt-tolerance mechanism: a review. Biochem Biophys Res Commun 495(1):286–291. https://doi.org/10.1016/j.bbrc.2017.11.043
Li QL, Xie JH, Ma XQ, Li D (2016) Molecular cloning of phosphoethanolamine N-methyltransferase (PEAMT) gene and its promoter from the halophyte Suaeda liaotungensis and their response to salt stress. Acta Physiol Plant 38(2):1–8. https://doi.org/10.1007/s11738-016-2063-4H
Li X, Li Y, Ahammed GJ et al (2019) RBOH1-dependent apoplastic H2O2 mediates epigallocatechin-3-gallate-induced abiotic stress tolerance in Solanum lycopersicum L. Environ Exp Bot 161:357–366. https://doi.org/10.1016/j.envexpbot.2018.11.013PLANTARUM
Luo D, Niu XL, Yu JD, Yan J, Gou XJ, Lu BR, Liu YS (2012) Rice choline monooxygenase (OsCMO) protein functions in enhancing glycine betaine biosynthesis in transgenic tobacco but does not accumulate in rice (Oryza sativa L. ssp. japonica). Plant Cell Rep 31:1625–1635. https://doi.org/10.1007/s00299-012-1276-2
Malekzadeh P (2015) Influence of exogenous application of glycinebetaine on antioxidative system and growth of salt-stressed soybean seedlings (Glycine max L.). Physiol Mol Biol Plants 21(2):225–232. https://doi.org/10.1007/s12298-015-0292-4
Mao Y, Chen N, Cao M, Chen R, Guan X, Wang D (2019) Functional characterization and evolutionary analysis of glycine-betaine biosynthesis pathway in red seaweed Pyropia yezoensis. Mar Drugs 17(1):70. https://doi.org/10.3390/md17010070
McNeil SD, Nuccio ML, Ziemak MJ, Hanson AD (2001) Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc Natl Acad Sci USA 98(17):10001–10005. https://doi.org/10.1073/pnas.171228998
McNeil SD, Rhodes D, Russell BL, Nuccio ML, Shachar-Hill Y, Hanson AD (2000) Metabolic modeling identifies key constraints on an engineered glycine betaine synthesis pathway in tobacco. Plant Physiol 124(1):153–162. https://doi.org/10.1104/pp.124.1.153
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nawaz M, Wang Z (2020) Abscisic acid and glycine betaine mediated tolerance mechanisms under drought stress and recovery in axonopus compressus: a new insight. Sci Rep 10(1):6942. https://doi.org/10.1038/s41598-020-63447-0
Nie Q, Qiao G, Peng L, Wen X (2019) Transcriptional activation of long terminal repeat retrotransposon sequences in the genome of pitaya under abiotic stress. Plant Physiol Biochem 135:460–468. https://doi.org/10.1016/j.plaphy.2018.11.014
Niu GL, Gou W, Han XL et al (2018a) Cloning and functional analysis of phosphoethanolamine methyltransferase promoter from maize (Zea mays L.). Int J Mol Sci 19(1):191. https://doi.org/10.3390/ijms19010191
Niu J, Zhang S, Liu S et al (2018b) The compensation effects of physiology and yield in cotton after drought Stress. J Plant Physiol 224–225:30–48. https://doi.org/10.1016/j.jplph.2018.03.001
Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD (1998) The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J 16(4):487–496. https://doi.org/10.1046/j.1365-313x.1998.00316.x
Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD (2000) cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in schizosac charomycespombe and characterization of the recombinant enzyme. J Biol Chem 275(19):14095–14101. https://doi.org/10.1074/jbc.275.19.14095
Ortiz TA, Takahashi LS (2015) Physical and chemical characteristics of pitaya fruits at physiological maturity. Genet Mol Res 14(4):14422–14439. https://doi.org/10.4238/2015.November.18.5
Raza SH, Athar HR, Ashraf M, Hameed A (2007) Glycine betaine induced modulation of antioxidant enzymes activities and ion accumulation in two wheat cultivars diffffering in salt tolerance. Environ Exp Bot 60(3):368–376. https://doi.org/10.1016/j.envexpbot.2006.12.009
Razavi F, Mahmoudi R, Rabiei V, Aghdam MS, Soleimani A (2018) Glycine betaine treatment attenuates chilling injury and maintains nutritional quality of hawthorn fruit during storage at low temperature. Sci Hortic 233:188–194. https://doi.org/10.1016/j.scienta.2018.01.053
Ren Z, Tang S, Jiang Y et al (2018) High-throughput sequencing analysis of endophytic bacteria diversity in fruits of white and red pitayas from three different origins. Pol J Microbiol 67(1):27–35. https://doi.org/10.5604/01.3001.0011.6139
Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol 44:357–384. https://doi.org/10.1146/annurev.pp.44.060193.002041
Scholz A, Stahl J, de Berardinis V, Müller V, Averhoff B (2016) Osmotic stress response in acinetobacter baylyi: Identifification of a glycine-betaine biosynthesis pathway and regulation of osmoadaptive choline uptake and glycine-betaine synthesis through a choline-responsive BetI repressor. Environ Microbiol Rep 8(2):316–322. https://doi.org/10.1111/1758-2229.12382
Tabuchi T, Kawaguchi Y, Azuma T, Nanmori T, Yasuda T (2005) Similar regulation patterns of choline monooxygenase, phosphoethanolamine N-methyltransferase and S-adenosyl-L-methionine synthetase in leaves of the halophyte Atriplex nummularia L. Plant Cell Physiol 46(3):505–513. https://doi.org/10.1093/pcp/pci050
Tabuchi T, Okada T, Takashima Y et al (2006) Transcriptional response of glycine betaine-related genes to salt stress and light in leaf beet. Plant Biotechnol 23(3):317–320. https://doi.org/10.5511/plantbiotechnology.23.317
Thirunavukkarasu N, Sharma R, Singh N et al (2017) Genomewide expression and functional interactions of genes under drought stress in maize. Int J Genomics 2017:2568706. https://doi.org/10.1155/2017/2568706
Tisarum R, Theerawitaya C, Samphumphuang T, Singh HP, Cha-Um S (2019) Foliar application of glycinebetaine regulates soluble sugars and modulates physiological adaptations in sweet potato (Ipomoea batatas) under water deficit. Protoplasma 257(1):197–211. https://doi.org/10.1007/s00709-019-01429-4
Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45(4):523–539. https://doi.org/10.1111/j.1365-313X.2005.02593.x
Wei D, Zhang W, Wang C et al (2017) Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. Plant Sci 257:74–83. https://doi.org/10.1016/j.plantsci.2017.01.012
Wu S, Yu Z, Wang F et al (2007) Cloning, characterization, and transformation of the phosphoethanolamine N-methyltransferase gene (ZmPEAMT) in maize (Zea mays L.). Mol Biotechnol 36(2):102–112. https://doi.org/10.1007/s12033-007-0009-1
Xu XY, Yao XZ, Lu LT et al (2018) Overexpression of the transcription factor NtNAC2 confers drought tolerance in tobacco. Plant Mol Biol Rep 36:543–552. https://doi.org/10.1007/s11105-018-1096-9
You L, Song Q, Wu Y, Li S, Jiang C, Chang L, Yang X, Zhang J (2019) Accumulation of glycine betaine in transplastomic potato plants expressing choline oxidase confers improved drought tolerance. Planta 249(6):1963–1975. https://doi.org/10.1007/s00425-019-03132-3
Yu J, Li Y, Tang W, Liu J, Lu BR, Liu Y (2014) The accumulation of glycine betaine is dependent on choline monooxygenase (oscmo), not on phosphoethanolaminen-methyltransferase (ospeamt1), in rice (Oryza sativa L. ssp. japonica). Plant Mol Biol Rep 32(4):916–922. https://doi.org/10.1007/s11105-014-0703-7
Zhang N, Si HJ, Wen G, Du HH, Liu BL, Wang D (2011) Enhanced drought and salinity tolerance in transgenic potato plants with a BADH gene from spinach. Plant Biotechnol Rep 5:71–77. https://doi.org/10.1007/s11816-010-0160-1
Zhang Y, Yin H, Li D, Zhu W, Li Q (2008) Functional analysis of BADH gene promoter from Suaeda liaotungensis K. Plant Cell Rep 27:585–592. https://doi.org/10.1007/s00299-007-0459-8
Zhang D, Tong J, He X et al (2016) A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic stress. Front Plant Sci 6:1237. https://doi.org/10.3389/fpls.2015.01237
Acknowledgements
This project is supported by grants from the National Natural Science Foundation of China (Grant No. 31760566) and the Innovation Talent Program of Guizhou Province, China (2016-4010).
Author information
Authors and Affiliations
Contributions
A-HW and X-PW conceived and designed the research. LY constructed the vector. X-ZY contributed to the bioinformatic analysis and obtained the transgenic tobacco plants. A-HW cloned HpPEAMT and performed all the experiments on the tobacco plants, analyzed the data, and prepared the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest to disclose.
Additional information
Communicated by Degao Liu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, AH., Yang, L., Yao, XZ. et al. Overexpression of the pitaya phosphoethanolamine N-methyltransferase gene (HpPEAMT) enhanced simulated drought stress in tobacco. Plant Cell Tiss Organ Cult 146, 29–40 (2021). https://doi.org/10.1007/s11240-021-02040-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02040-3