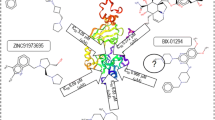
Abstract
The Ebola virus is a deadly pathogen that causes a highly lethal hemorrhagic fever illness in humans, sometimes known as Ebola virus sickness (EVD). The Ebola virus polymerase cofactor VP35 acts by preventing the establishment of a cellular antiviral state by blocking virus-induced phosphorylation and activation of interferon regulatory factor 3 (IRF3), a transcription factor required for the induction of interferons alpha and beta, thus making it an appealing therapeutic target because there are currently not many available and effective therapeutic agents available against this virus. This study presented a molecular docking–based virtual screening (VS) of 10,829 compounds acquired from multiple databases against the VP35 receptor using Auto Dock Vina software to discover potential inhibitors. According to the results of the screening, the top two drugs, irinotecan and fexofenadine, exhibited a high affinity for the VP35 binding region. Their binding affinities were −8.2 and −8.0 kJ/mol, indicating that they were tightly bound to the target receptor. These results outperformed those obtained with the co-crystallized ligand, which exhibited a binding affinity of −6.8 kJ/mol. As a result of the VS and molecular docking techniques, novel VP35 inhibitors from diverse databases were discovered using the Lipinski rule of five and functional molecular interactions with the target protein, as proven by the findings of this work. The findings suggest that the compounds discovered may offer viable avenues for the development of Ebola virus VP35 inhibitors and that they need further evaluation and investigation.









Similar content being viewed by others
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Code availability
Not applicable.
References
Weyer J, Grobbelaar A, Blumberg L (2015) Ebola virus disease: history, epidemiology, and Outbreaks. Curr Infect Dis Rep 17(5):480
Brown CS, Lee MS, Leung DW, Wang T, Xu W, Luthra P et al (2014) In silico derived small molecules bind the filovirus VP35 protein and inhibit its polymerase cofactor activity. J Mol Biol 426(10):2045–2058
Mohamad Yusoff MA, Abdul Hamid AA, Mohammad Bunori N, Abd Halim KB (2018) Interaction of monomeric Ebola VP40 protein with a plasma membrane: a coarse-grained molecular dynamics (CGMD) Simulation Study. J Mol Graph Model 82:137–144. https://doi.org/10.1016/j.jmgm.2018.04.010
Ahmad N, Farman A, Badshah SL, Ur Rahman A, ur Rashid H, Khan K (2017) Molecular modeling, simulation and Docking Study of Ebola virus glycoprotein. J Mol Graph Model 72:266–71. https://doi.org/10.1016/j.jmgm.2016.12.010
Holmes EC, Dudas G, Rambaut A, Andersen KG (2016) The evolution of ebola virus: insights from the 2013–2016 epidemic. Nature 538(7624):193–200. https://doi.org/10.1038/nature19790
Leung DW, Prins KC, Basler CF, Amarasinghe GK (2010) Ebolavirus VP35 is a multifunctional virulence factor. Virulence 1(6):526–31. https://doi.org/10.4161/viru.1.6.12984
Cárdenas WB, Loo YM, Gale Jr M, Hartman AL, Kimberlin CR, Martínez-Sobrido L et al (2006) Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by rig-I signaling. J Virol 80(11):5168–78. https://doi.org/10.1128/JVI.02199-05
Sliwoski G, Kothiwale S, Meiler J, Lowe EW (2013) Computational methods in drug discovery. Pharmacol Rev 66(1):334–95. https://doi.org/10.1124/pr.112.007336
Taylor NR, Smith R (1996) The World Wide Web as a graphical user interface to program macros for molecular graphics, molecular modeling, and structure-based drug design. J Mol Graph 14(5):291–296. https://doi.org/10.1016/S0263-7855(96)00077-X
Chen H, Zhou X, Gao Y, Chen H, Zhou J (2017) Fragment-based drug design: strategic advances and lessons learned. Comprehensive Med Chem III:212–232. https://doi.org/10.1016/B978-0-12-409547-2.12319-4
Prashantha CN, Gouthami K, Lavanya L, Bhavanam S, Jakhar A, Shakthiraju RG et al (2021) Molecular screening of antimalarial, antiviral, anti-inflammatory and HIV protease inhibitors against Spike glycoprotein of coronavirus. J Mol Graph Model 102:107769. https://doi.org/10.1016/j.jmgm.2020.107769
Zhang M-Z, Chen Q, Yang G-F (2015) A review on recent developments of indole-containing antiviral agents. Eur J Med Chem 89:421–41. https://doi.org/10.1016/j.ejmech.2014.10.065
Han C, Guo Y-C, Wang D-D, Dai X-J, Wu F-J, Liu H-F et al (2015) Novel pyrazole fused heterocyclic ligands: synthesis, characterization, DNA binding/cleavage activity and anti-BVDV activity. Chin Chem Lett 26(5):534–38. https://doi.org/10.1016/j.cclet.2015.01.006
Shaker YM, Omar MA, Mahmoud K, Elhallouty SM, El-Senousy WM, Ali M et al (2015) Synthesis, in vitro and in vivo antitumor and antiviral activity of novel 1-substituted benzimidazole derivatives. J Enzyme Inhib Med Chem 30(5):826–845. https://doi.org/10.3109/14756366.2014.979344
Gueiffier A, Mavel S, Lhassani M, Elhakmaoui A, Snoeck R, Andrei G et al (1998) Synthesis of imidazo[1,2-a]pyridines as antiviral agents. J Med Chem 41(25):5108–5112. https://doi.org/10.1021/jm981051y
Srivastava PC, Pickering MV, Allen LB, Streeter DG, Campbell MT, Witkowski JT et al (1977) Synthesis and antiviral activity of certain thiazole C-nucleosides. J Med Chem 20(2):256–262. https://doi.org/10.1021/jm00212a014
Albratty M, El-Sharkawy KA, Alhazmi HA (2019) Synthesis and evaluation of some new 1,3,4-oxadiazoles bearing thiophene, thiazole, coumarin, pyridine and pyridazine derivatives as antiviral agents. Acta Pharm 69(2):261–76. https://doi.org/10.2478/acph-2019-0015
Ali EMH, Abdel-Maksoud MS, Oh C-H (2019) Thieno[2,3-d]pyrimidine as a promising scaffold in medicinal chemistry: recent advances. Bioorg Med Chem 27(7):1159–1194. https://doi.org/10.1016/j.bmc.2019.02.044
Wei Y, Wang H, Xi C, Li N, Li D, Yao C et al (2020) Antiviral effects of novel 2-benzoxyl-phenylpyridine derivatives. Molecules 25(6):1409. https://doi.org/10.3390/molecules25061409
Trott O, Olson A (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Visualizer DS (2005). v4. 0.100. 13345. Accelrys Software Inc, 2013
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Lee SK, Lee IH, Kim HJ, Chang GS, Chung JE, No KT (2003) The PreADME approach: web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties. EuroQSAR 2002 Designing Drugs and Crop Protectants: processes, problems and solutions 418–20
Baell J, Ferrins L, Falk H, Nikolakopoulos G (2013) PAINS: relevance to tool compound discovery and fragment-based screening. Aust J Chem 66(12):1483–1494. https://doi.org/10.1071/CH13551
Daina A, Zoete V (2016) A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem Med Chem 11(11):1117–1121. https://doi.org/10.1002/cmdc.201600182
López-Blanco J, Aliaga J, Quintana-Ortí E, Chacón P (2014) iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 42(W1):W271–W276. https://doi.org/10.1093/nar/gku339
Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. InSC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE, pp 43–43
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang J, Wang L, Lupyan D, Dahlgren M, Knight J, Kaus J, Cerutti D, Krilov G, Jorgensen W, Abel R, Friesner R (2015) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. https://doi.org/10.1021/acs.jctc.5b00864
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6:1509–1519. https://doi.org/10.1021/ct900587b
López-Blanco J, Aliaga J, Quintana-Ortí E, Chacón P (2014) iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 42:W271–W276. https://doi.org/10.1093/nar/gku339
Acknowledgements
The authors would like to acknowledge the Bioinformatics lab facility of the School of Interdisciplinary Studies, Jamia Hamdard University New Delhi, 110062, India, during the work. The help from Dr. Mymoona Akhter, Bioinformatics lab, is also of great importance, particularly during the MD simulation analysis. We also thank BioNome that arranged the Desmond Schrodinger part in the manuscript for simulation studies.
Author information
Authors and Affiliations
Contributions
Ratul Bhowmik conducted database searches, compiled references, evaluated data, and wrote the article; Ajay Manaithiya contributed significantly to the design, analysis, manuscript preparation, revision, and writing—original draft; Bharti Vyas contributed to the conception of the study and formal analysis; Ranajit Nath helped to organize the literature; Sara Rehman contributed to the collection of data and checked the chemical structures; Shubham Roy polished the language; Ratna Roy contributed to the table and picture layout. All authors contributed to the paper. All authors read and approved the final manuscript. They warrant that the article is the authors’ original work, has not received prior publication, and is not under consideration for publication elsewhere.
Corresponding author
Ethics declarations
Ethical approval
All the co-authors are aware of and approve of the submission.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• A structural library including 10829 ligands was evaluated via molecular docking against the Ebola virus VP35.
• Fexofenadine and irinotecan have a high affinity for Ebola virus VP35 when used as a protein-ligand complex.
• IMODs and Desmond were used to determine the flexibility and disordered areas of proteins.
• Selected compounds have been shown to inhibit the Ebola virus VP35.
• Virtual high-throughput screening and pharmacoinformatics aid in the development of new drugs.
Rights and permissions
About this article
Cite this article
Bhowmik, R., Manaithiya, A., Vyas, B. et al. Identification of potential inhibitor against Ebola virus VP35: insight into virtual screening, pharmacoinformatics profiling, and molecular dynamic studies. Struct Chem 33, 815–831 (2022). https://doi.org/10.1007/s11224-022-01899-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-022-01899-y