Abstract
The potential energy surfaces of stacked structures consisting of adenine (A) and 2-aminopurine (2AP) have been investigated in the gas phase. Both face-to-back (the double-ring system of one base exactly on top of that of the other one) and face-to-face (one base flipped by 180°) A/A, 2AP/2AP and A/2AP stacks were considered. Minima and transition states were optimised at the counterpoise-corrected M06-2X/6-31+G(d) level of theory. For each type of stack, between five and nine minima were located, usually connected by low barriers of 1–2 kcal/mol. This shows the large conformational flexibility of these stacked base pairs. The double-ring system in A and 2AP affords multiple minima with similar twist angles, making the potential energy surface of stacks comprising of purine bases more complex than those of pyrimidine stacks. The locations of the stationary points on the potential energy surface differ for the three different systems; thus, the replacement of A by 2AP in a base stack changes its potential energy landscape.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The stability of the DNA and RNA double helix arises from a fine balance of hydrogen (H-) bonding interactions between complementary bases on opposite strands (termed Watson–Crick base pairing), H-bonding interactions between the bases and surrounding water molecules, as well as π-stacking interactions between adjacent bases on one strand. Even though these interactions tend to be weaker than H-bonds in terms of potential energy, it is now well recognised that π-stacking is more important than base pairing as a thermodynamic driving force for the stabilisation of DNA [1–3]. A comprehensive understanding of base stacking is therefore paramount for understanding DNA stability. In earlier work, we studied the potential energy surfaces of stacked uracil dimer (U/U), stacked thymine dimer (T/T) and stacked 5-bromouracil–uracil dimer (5BrU/U) [4, 5]. Potential energy scans were created by rotating one of the bases in the stack while keeping the lower molecule fixed. Minima and transition states thus identified were subsequently fully optimised with M06-2X/6-31+G(d). Several minima with varying twist angles were identified for each system, often separated by low barriers. These results provide further structural basis for our understanding of the torsional flexibility of DNA structures [6–8]. Comparison of the M06-2X/6-31+G(d) interaction energies of optimised minimum-energy structures of U/U stacks with estimated CCSD(T)/CBS (complete basis set) results revealed the excellent performance of this functional/basis set combination for stacking energies. In the current work, the same methodology is applied to stacks of the purine base adenine (A) and its analogue 2-aminopurine (2AP). Our interest in 2AP comes from the fact that it is readily incorporated into DNA in place of adenine and does not significantly alter DNA structure. Unlike adenine, 2AP is fluorescent and this property has been used for many years to experimentally investigate local nucleic acid structure [9–14]. As a first step, we are interested in investigating whether 2AP behaves differently from A in base stacking. In addition, we will investigate whether stacks consisting of purines have more flexibility (more local minima) compared with stacks consisting of pyrimidines, as the double-ring system in purines may offer more possibilities of favourable π–π interactions.
Above, we mentioned the excellent performance of the M06-2X/6-31+G(d) level of theory for DNA base stacking. An additional justification of using M06-2X (rather than higher-level ab initio methods) in this work is the ability of DFT-based methods to study larger DNA structures. DFT calculations on large DNA fragments have started to appear in the literature. For example, Šponer et al. [15] used dispersion-corrected DFT (DFT-D3) to study two-quartet G-DNA stems, whereas Shishkin, Leszczynski et al. used the M06-2X functional to study double-stranded mini-helices [16]. For these systems, modern DFT methods offer much better description than semi-empirical or molecular mechanics methods.
The first electronic structure study on A/A was reported in 1996 [17]. Since then, many studies incorporating calculations on A/A have appeared in the literature [7, 17–29]. Most of these use geometries based on either crystal structure data [7, 19, 20, 25, 28] or structures consisting of rigid bases [17, 18, 21, 23, 24, 26, 29]. Waller et al. [22] fully optimised an A/A stack with the BH&H functional, but its structure is not reported. Wang [27] fully optimised five A/A stacks at the MPWB1K/cc-pVDZ level of theory. Most studies on A/A stacks used some variant of MP2 or DFT to calculate the stacking energies. A few studies provide reference CCSD(T) data. Extrapolated complete basis set (CBS) interaction energies at the CCSD(T) level were reported for base stacking in B-DNA base pair steps [21] and for 131 structures of A/A [29]. These studies show that, at the CBS limit, MP2 overestimates the interaction. The latter study also investigated the performance of the M06-2X functional for stacked A/A structures and found that it performed well.
DNA bases have two distinguishable faces, and isolated base stacks can therefore associate in different ways. Rose et al. [30] suggested an α/β-face nomenclature based on the clockwise or anticlockwise direction of ascending atom numbering in the ring with the lowest-numbered unshared ring atom. If progression from the lowest-numbered to the next higher-numbered atom (by the shortest route) is in the clockwise direction, then the face is α; if the progression is anticlockwise, it is β. The bases on the same strand of a double helix with antiparallel strands usually face in the same direction. When looking down a strand from the 5′ towards the 3′-end, the purines will show their β-faces and the pyrimidines their α-faces [30]. Thus, adjacent purines stack with the α-side of the 5′-base against the β-side of the base at the 3′-end [31]. This is also referred to as face-to-back stacking [32]. Face-to-face stacking occurs in cross-strand stacking interactions. In this work, both face-to-back and face-to-face arrangements are considered.
Methodology
The structures of the A and 2AP bases were optimised using the M06-2X [33] density functional and the 6-31+G(d) basis set. The 9H-tautomer (see Scheme 1) was used for both A and 2AP, which is the most stable form for both bases [34–36]. Stacked A/A, A/2AP and 2AP/2AP structures were built by placing the two molecules in a parallel arrangement. Both face-to-face and face-to-back structures were considered for each base pair combination. In the face-to-back structures, the ring systems of the two molecules are placed exactly on top of each other (see Scheme 1). In the face-to-face structures, the upper molecule is flipped by 180° (rotation of 180° around the C8-X axis). Dummy atoms (X in Scheme 1) were placed in the midpoint of the C–C bond linking the five- and six-membered rings. The inter-base distance (as measured by the distance between the two dummy atoms) was set at 3.4 Å (the approximate distance between consecutive bases in B-DNA [37]). In the nomenclature of Rose et al. [30], face-to-back stacking is referred to as β-α stacking, whereas face-to-face is referred to as β-β.
Rigid potential energy scans were created by rotating the upper molecule in the counterclockwise direction, while keeping the lower molecule fixed, by varying the C4(upper)-X(upper)-X(lower)-C4(lower) (for the face-to-back structures) or C4(upper)-X(upper)-X(lower)-C5(lower) torsion angle (for the face-to-face structures) in steps of 1°. Note that with this definition of the torsion angle (τtwist), for both face-to-back and face-to-face structures shown in Scheme 1, τtwist = 0°. Interaction energies, corrected for BSSE using the counterpoise (CP) [38] scheme, were calculated (see below) at each point of the scan.
Unlike in H-bonded DNA base pairs, where the amino groups are planar to optimise hydrogen bonding, in isolated gas-phase bases the amino groups show asymmetric pyramidalisation [39]. The potential energy scans therefore depend on whether the amino group hydrogens are in the “up” or “down” position. For A/A and 2AP/2AP, there are three different possibilities: down-up (du—where the first letter refers to the “lower” base and the second one to the “upper” base; see Scheme 1), ud, dd and uu. The ud and du face-to-back profiles of A/A and 2AP/2AP are symmetric around 180°; a stack with τtwist = τ° is symmetry-equivalent to a stack with τtwist = 360 − τ°. For these systems, dd and uu configurations with the same twist angle are mirror images of each other. For the face-to-face A/A and 2AP/2AP stacks, uu and dd configurations with the same twist angle are identical (they can be rotated into each other). For both face-to-back and face-to-face A/2AP, the four possibilities for the amino hydrogen positions (dd, du, ud and uu) yield distinct structures.
Full geometry optimisations were performed starting from the minima in the potential energy scans using M06-2X/6-31+G(d). Transition state optimisations were performed using the QSTN [40, 41] (N = 2 or 3) algorithm. The geometry optimisations were carried out on the counterpoise (CP)-corrected potential energy surfaces. CP-corrected interaction energies were calculated according to Eq. (1):
where the monomer deformation energies follow from:
(with a similar expression for E defB2 ). In Eqs. (1) and (2), B1 and B2 are the two bases in the stack; the superscripts in curly brackets indicate the basis set employed in the calculation (dimer basis set {B1/B2} or monomer basis set {B1} or {B2}); the subscripts indicate the molecular system; and the geometry (optimised stack geometry B1/B2 or monomer geometries B1 or B2) is indicated in round brackets. The CP-corrected interaction energies in the rigid scans were calculated in the same way. Note, however, that in the scans the monomer deformation energies are zero (and can thus be ignored), because we used M06-2X/6-31+G(d)-optimised monomer structures in the scans. Harmonic vibrational frequencies were calculated for the fully optimised structures at the same level of theory to verify the nature of the stationary points (minima or first-order saddle points) and to compute zero-point energy corrected interaction energies (denoted as ΔE CP0 ). All calculations were done with Gaussian 09 [42] and employed the “ultrafine” integration grid.
Several geometric parameters were calculated to characterise the stacked structures, including the rise (vertical distance between the two bases), slide (horizontal displacement) and tilt (angle between the planes of the bases)—see Scheme 1. These are based on the base pair step parameters introduced by Dickerson [43]. Note that for stacks of single bases (as opposed to stacks of base pairs) there is no distinction between slide and shift or between roll and twist. The rise is calculated as the scalar product of the vector connecting the midpoints of the central C–C bonds (m 1 m →2 ) and the plane vector of one of the bases (where a plane is defined by the C2, C4 and C5 atoms). As the two bases are not completely parallel, this scalar product depends on which base’s plane vector is used. The rise was therefore computed as the average of the two values obtained when using the first or the second base’s plane vector. The slide is calculated as the component of the vector connecting the two midpoints that is in the plane of one base. Like for the rise, the slide is calculated as the average of the two values obtained when using the first or the second base’s plane vector. The tilt is the angle between the plane vectors of the two bases.
Results
Face-to-back structures
The potential energy scans of the face-to-back A/A, A/2AP and 2AP/2AP stacks are shown in Fig. 1. All profiles exhibit a high maximum at τtwist = 0°. This is as expected, because at 0° the ring atoms of the upper base are exactly on top of the corresponding atoms of the lower base. The A/A and 2AP/2AP profiles are not exactly symmetric due to the pyramidality of the amino groups, which in the scan are oriented in the same direction (dd). We investigated the effect of the amino group orientation on the potential energy profiles (Fig. S1 in electronic supplementary material). Perfectly symmetric face-to-back A/A and 2AP/2AP profiles are obtained when the amino hydrogens are in ud or du orientation. For these, every structure with twist angle τ° has a mirror image at 360° − τ°. A dd A/A or 2AP/2AP structure with twist angle τ° has a mirror image that is a uu stack with τtwist = 360° − τ°. The 2AP/2AP dd profile deviates much more from symmetry than the corresponding A/A profile, because in the optimised 2AP monomer the amino protons are further out of the plane of the ring system as compared to A. The face-to-back A/2AP profiles do not exhibit any symmetry. The four A/2AP profiles clearly fall into two groups: du and uu in one group and dd and ud in the other group. Apparently, the orientation of the upper base, 2AP, is more important for determining the shape of the profile. Again, this is presumably due to the greater non-planarity of the amino group in 2AP compared with A. The twist angle dependence of the A/A energy has been investigated before. Šponer et al. [17] created scans of the twist angle using MP2/6-31G*(0.25). Elstner et al. [18] used SCC-DFTB augmented with a dispersion term as well as MP2/6-31G*(0.25). Ducéré and Cavallo [23] used DFT with the PBE functional augmented with a dispersion term as well as MP2/aug-cc-pVTZ. Morgado et al. [29] calculated scans using a range of methods, including MP2/CBS corrected for higher-order electron correlation using the CCSD(T) method. Most of these (except the SCC-DFTB profiles) show a maximum at about 140°–150° instead of the maximum at 180° in our scans. However, our fully optimised structures show that the A/A stack with twist angle 180° is a transition state (see below and Table 1a), in accordance with our profiles.
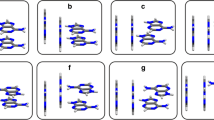
Geometry optimisations were started from the minima in the profiles. In some cases (notably transition states where the imaginary frequency mode did not resemble a simple twist angle rotation), geometry optimisations were initiated from structures generated from the transition state moved slightly along both directions of the reaction path (by adding or subtracting the Cartesian coordinates of the displacements of the nuclei associated with the imaginary frequency mode). Below we refer to this as the “following the imaginary frequency” method. The optimised minima and transition states are displayed in Figs. 2, 3 and 4. Cartesian coordinates of all face-to-back minima and transition states are also provided as electronic supplementary material (Table S1). Geometric and energetic characteristics are listed in Table 1.
The A/A global minimum occurs at a twist angle of 136° (Table 1a). Note that there is a mirror image of the 136°-minimum at 224°. The transition state linking these symmetry-related minima (at τtwist = 180°) is Ci-symmetric with perfectly parallel bases (tilt = 0°). There are further minima at twist angles of 3°, 58° and 62°. The 19°-transition state presumably links the minima at 3° and 58°, whereas the 83°-transition state presumably links the minima at 62° and 136° (and similarly for their symmetry-related equivalents). The 19°-transition state exhibits a large slide (1.65 Å), to avoid atoms in the upper base to be stacked directly on top of the corresponding atoms in the lower base. The rise is also larger than in the other A/A structures, reflecting the less favourable interaction energy in this transition state. The transition state at 59° was obtained with the minima at 58° and 62° as reactant and product structures in the QST2 input. It has a similarly large slide as the 58°-minimum, but a larger tilt. As noted above, the amino groups can have different relative orientations (u or d). For the 62°-minimum, all different amino group positions converged to the ud isomer listed in Table 1a. We have not systematically examined different amino group orientations for the other A/A minima and transition states. For 2AP/2AP several pairs of minima were located that mainly differ in the amino group orientations (vide infra). However, it is expected that the amino groups can readily interconvert between different orientations. Wang located three face-to-back A/A structures by varying the relative orientations of the five- and six-membered rings [27]. The structures were fully optimised at the MPW1K/cc-pVDZ level of density functional theory. Wang does not report geometrical details, but inspection of the structures displayed in Fig. 1 of Ref. [27] indicates that the “displaced parallel,” “displaced antiparallel” and “rotated” stacks are similar to those with twist angles 3°, 136° and 58°/62° located in the current work, respectively. Wang calculates single-point MP2/6-311++G(d,p) interaction energies of −7.7, −10.7 and −9.1 kcal/mol for these three structures, in close agreement with our M06-2X/6-31+G(d) results of −7.45, −10.43 and −8.97 kcal/mol for stacks with twist angles of 3°, 136° and 62°, respectively. Note that at the CBS limit, MP2 overestimates the stacking energy [21, 29]. However, the basis set incompleteness error in MP2/6-311++G(d,p) likely cancels the overestimation to some extent. Šponer et al. [21] reported an extrapolated CCSD(T)/CBS interaction energy of −8.5 kcal/mol for an A/A structure taken from earlier work [17]. This is a non-optimised structure. With twist angle around 120°, rise of 3.3 Å and 0° tilt, it is close to the global minimum at τtwist = 136°, for which an interaction energy of −10.43 kcal/mol was calculated. As the structure of Šponer et al. is not optimised, the interaction energy will not be optimal; a difference of 2 kcal/mol between the CCSD(T) and M06-2X interaction energies therefore seems very plausible. We are not aware of a CCSD(T) reference value for a fully optimised A/A stack.
For 2AP/2AP only the symmetry-unique stationary points are listed in Table 1b. The global minimum occurs at τtwist = 180° (ΔE CP = 9.96 kcal/mol). This one has the amino groups in the du orientation. Another minimum with τtwist = 180° and ud amino hydrogens is more than 1 kcal/mol less stable. Attempts to locate a minimum with twist angle 180° and amino hydrogens in the uu (or dd) position led to the minimum with τtwist = 161° listed in Table 1b. This is presumably an intermediate between the 180-ud and 180-du minima. Transition-state optimisations with the 161°-minimum as the reactant and either the 180-du or 180-ud minima as the product structure in the QST2 input led to transition states with twist angles of 169° and 175°, respectively. Also for some other minima two different arrangements of the amino hydrogens were found. For example, for τtwist = 61°/62°, there are two distinct minima, one with ud and one with dd orientation. A transition state with pd (where p indicates an essentially planar amino group) connects these two minima. Note that the barrier for conversion between the two minima is very low (~0.3 kcal/mol). A geometry optimisation with the amino groups in the uu orientation converged to the ud minimum, whereas a geometry optimisation starting from du converged to the dd minimum. Likewise, there are two minima with τtwist = 101°/102° and 130°/133°. Both pairs are connected by a transition state with one planar amino group. We have not extensively investigated all possible amino group orientations for all minima and transition states. Presumably some of the A/A minima may also exist with differing amino group positions. Note, however, that the barriers for interconversion between different amino group orientations are small, as shown for the τtwist = 61°/62° and 130°/133° pairs, and it is expected that under physiological conditions the different stacks can convert freely into their corresponding forms. There likely exist more minima and transition states than those listed in Table 1b. It is clear that the 2AP/2AP potential energy surface contains many minima, with mostly low barriers in between.
A recent two-dimensional fluorescence spectroscopic study on the aqueous-phase conformation of a dinucleotide containing 2AP as its bases found that the average conformation contains nearly eclipsed bases (twist angle of 5°) [44]. This contrasts with the gas-phase results obtained here, where the lowest energy structure has a twist angle of 180° and the stationary point closest to a twist angle of 0° is a transition state (τtwist = 27°). This is actually the stationary point with the smallest interaction energy (−6.50 kcal/mol). Geometry optimisation of a 2AP dinucleotide in the gas phase yielded a twist angle of 63° (calculated in the same way as in the current work) [45], which is very close to our minimum at 62° (ΔE CP = −9.24 kcal/mol). The disagreement with Ref. [44] could in principle be due to the aqueous environment in the fluorescence spectroscopy study. However, from our earlier work on stacking of the pyrimidines U, 5BrU and T [4, 5], face-to-back structures with twist angles close to 0° tend to be transition states with a relatively small interaction energy, and the inclusion of water via the continuum solvation model did not change this.
Also the face-to-back A/2AP potential energy surface contains many minima (Table 1c). We located 16 stationary points (9 minima and 7 transition states), but assume that there are more. The global minimum is at τtwist = 298° (with amino hydrogens in the dd position). A corresponding minimum (τtwist = 298°) with ud amino hydrogens is very close in energy and is separated from the dd minimum by a very shallow barrier (0.02 kcal/mol). There is a transition state (τtwist = 352°; ΔE CP = −6.47 kcal/mol) near the 0°-maximum in the scan.
Face-to-face structures
The potential energy scans of the face-to-face A/A, A/2AP and 2AP/2AP stacks (with the amino hydrogens in the dd position) are shown in Fig. 5. In contrast to the face-to-back profiles, the face-to-face potential energy curves do not show a prominent maximum at τtwist = 0°; only the 2AP/2AP face-to-face profile has a low maximum at 0°. The profiles with du and ud (and uu for A/2AP) are shown in supplementary material (Fig. S1). The 2AP/2AP profiles differ most from one another, presumably due to the large non-planarity of the amino groups. Like for the face-to-back profiles, the upper base, 2AP, predominantly determines the shape of the face-to-face A/2AP profiles.
As for the face-to-back structures, geometry optimisations were started from the minima in the profiles, and transition states were located using the QST2 and QST3 methods, whereas in some cases additional minima were located by the “following the imaginary frequency” method. The optimised minima and transition states are displayed in Figs. 6, 7 and 8. Cartesian coordinates of all face-to-face minima and transition states are also provided as electronic supplementary material (Table S2). Geometric and energetic characteristics are listed in Table 2.
Table 2a lists the located minima and transition states of the face-to-face A/A stacked dimer. We found minima at twist angles of 7°, 100°, 161°, 239°, 243° and 310°. These roughly correspond to the troughs in the profile displayed in Fig. 5, except for the minimum at 7°, where the profile does not show a minimum, and the minima at 239° and 243°, where the profile shows a maximum. A transition state with planar amino group for the lower base connects these latter two minima. The lowest energy minimum occurs at τtwist = 161°. We did not locate a transition state between the minima with twist angles of 161° and 243°.
In addition to the three face-to-back A/A stacks discussed above, Wang also located two face-to-face A/A structures using MPW1K/cc-pVDZ [27]. These appear to be similar to our face-to-face stack with twist angle 7°. The MP2/6-311++G(d,p) single-point interaction energy of one of Wang’s structures (−9.5 kcal/mol) is very close to the M06-2X/6-31+G(d) interaction energy of our 7°-structure (−9.45 kcal/mol). Wang did not locate a structure similar to our 161°-minimum, which is the lowest-energy face-to-face A/A stack we found. This confirms that locating all A/A structures is not a simple task.
Table 2b lists the stationary points found for the face-to-face 2AP/2AP system. Again, the minima roughly correspond to the troughs in the profile in Fig. 5. The minima at 6 and 9° occur at smaller twist angle than predicted by the profile, and a transition state was found at 310° instead of 0/360°. The 6°- and 9°-minima mainly differ in the amino group orientation (ud for the 6°-minimum and dd for the 9°-minimum). A transition state (τtwist = 8°) with nearly planar amino group in one of the 2AP bases connects these two minima. We did not find a minimum around 350°, as shown in the profile. The lowest-energy minimum occurs at τtwist = 154°.
Table 2c lists the stationary points for the face-to-face A/2AP stacked dimer. The global minimum is a structure with τtwist = 169°. Several minima and transition states in the τtwist = 0°–30° range were found, some differing by only 1° in the twist angle. The 14°-transition state was obtained by a QST2 optimisation with the 6°- and 13°-minima as reactant and product geometries, whereas the 15°-transition state resulted from using the 13°- and 17°-minima in the QST2 input. The minima at 13 and 17° mainly differ in the direction the amino protons of the lower base (A) are pointing (towards or away from the upper base). The minima at 159 and 169° principally differ in the direction of the slide.
Discussion and conclusions
The potential energy surfaces of face-to-back and face-to-face A/A, 2AP/2AP and A/2AP stacks were explored at the M06-2X/6-31+G(d) level of theory. We used the same methodology as previously employed for stacks comprising the pyrimidine bases U, 5BrU and T [4, 5], which involves first creating rigid potential energy curves by rotating one of the bases in the stack while keeping the other one fixed. The minima in the curves were then used as starting structures for geometry optimisations.
For all different base stacks, a number of different minima were found. Some differ mainly in the orientation of the amino hydrogens, which can be “up” or “down” for each individual base in the stack. The rise is generally 3.1–3.2 Å, somewhat smaller than the average value of 3.4 Å in B-DNA [37]. The presence of the double-ring system in the purines means that different minima may exist with similar twist angle but different slides. For example, the face-to-back A/2AP minima with twist angles 56° and 59° differ mainly in the slide (note that our definition of slide does not include direction). This indicates that our methodology for locating minima by rotation of one base (i.e. variation of just the twist angle) cannot be guaranteed to find all minima. A more comprehensive search algorithm would involve simultaneously scanning twist and slide parameters (slide in several directions). In a number of cases, additional minima were found by “following the imaginary frequency” of a transition state (in both directions). Overall, the stacks including A and 2AP display more complex potential energy surfaces containing more minima, as compared to the pyrimidine stacks comprising U, 5BrU and T studied previously [4, 5]. Minima and transition states occur at different twist, slide and tilt values for A/A, 2AP/2AP and A/2AP. Thus, the replacement of A by 2AP changes the energy landscape of the stack.
Unlike in the isolated stacks presented in this work, in the DNA duplex the bases cannot rotate freely, as they are restricted by the sugar–phosphate backbone. In an idealised DNA helix, the twist is 36°. However, it is known that DNA is highly dynamic and that its structure can both bend and twist [8]. The first right-handed DNA oligomer that was resolved by single-crystal X-ray crystallography showed sequence-dependent variations of the step parameters, including the twist angle, which varied from 32° to 45° [46]. By analysis of a database consisting of 40 DNA oligomers, El Hassan and Calladine showed that the twist can vary from as little as 10° to as much as 60° [37]. The slide (or shift) typically varies from −3 to +3 Å, whereas the tilt (or roll) can vary between −25° to +25°. The A/A, 2AP/2AP and A/2AP face-to-back structures, which are most relevant to double-helical DNA, all have a minimum around τtwist = 60°, at the border of what can be achieved in DNA. In all cases, this is not the most stable minimum. This shows that the stacking configuration in DNA is not optimal in terms of pure base/base stacking. However, our results also show that the potential energy surfaces of the base stacks are rather flat, with barriers typically less than 3 kcal/mol. This indicates that, in terms of base/base stacking, it would not cost much energy to rotate a base from its optimal position in an isolated stack.
References
Bommarito S, Peyret N, SantaLucia J (2000) Thermodynamic parameters for DNA sequences with dangling ends. Nucl Acids Res 28:1929–1934
Kool ET (2001) Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu Rev Biophys Biomol Struct 30:1–22
Yakovchuk P, Protozanova E, Frank-Kamenetskii MD (2006) Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 34:564–574
Hunter RS, van Mourik T (2012) DNA base stacking: the stacked uracil/uracil and thymine/thymine minima. J Comput Chem 33:2161–2172
Holroyd LF, van Mourik T (2014) Stacking of the mutagenic DNA base analog 5-bromouracil. Theor Chem Acc 133:1–13
Hagerman PJ (1988) Flexibility of DNA. Ann Rev Biophys Chem 17:265–286
Samanta S, Kabir M, Sanyal B, Bhattacharyya D (2008) Twist-dependent stacking energy of base-pair steps in B-DNA geometry: a density functional theory approach. Int J Quant Chem 108:1173–1180
Travers AA (2004) The structural basis of DNA flexibility. Phil Trans R Soc Lond A 362:1423–1438
Stivers JT (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucl Acids Res 26:3837–3844
Rachofsky EL, Seibert E, Stivers JT, Osman R, Ross JBA (2001) Conformation and dynamics of abasic sites in DNA investigated by time-resolved fluorescence of 2-aminopurine†. Biochemistry 40:957–967
Rai P, Cole TD, Thompson E, Millar DP, Linn S (2003) Steady-state and time-resolved fluorescence studies indicate an unusual conformation of 2-aminopurine within ATAT and TATA duplex DNA sequences. Nucl Acids Res 31:2323–2332
Jean JM, Hall KB (2004) Stacking–unstacking dynamics of oligodeoxynucleotide trimers†. Biochemistry 43:10277–10284
Johnson NP, Baase WA, von Hippel PH (2004) Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA. Proc Natl Acad Sci USA 101:3426–3431
Bonnist EYM, Jones AC (2008) Long-wavelength fluorescence from 2-aminopurine-nucleobase dimers in DNA. Chem Phys Chem 9:1121–1129
Šponer J, Mládek A, Špačková N, Cang X, Cheatham TE, Grimme S (2013) Relative stability of different DNA guanine quadruplex stem topologies derived using large-scale quantum-chemical computations. J Am Chem Soc 135:9785–9796
Zubatiuk TA, Shishkin OV, Gorb L, Hovorun DM, Leszczynski J (2013) B-DNA characteristics are preserved in double stranded d(A)3·d(T)3 and d(G)3·d(C)3 mini-helixes: conclusions from DFT/M06-2X study. Phys Chem Chem Phys 15:18155–18166
Šponer J, Leszczynski J, Hobza P (1996) Nature of nucleic acid-base stacking: nonempirical ab initio and empirical potential characterization of 10 stacked base dimers. Comparison of stacked and H-bonded base pairs. J Phys Chem 100:5590–5596
Elstner M, Hobza P, Frauenheim T, Suhai S, Kaxiras E (2001) Hydrogen bonding and stacking interactions of nucleic acid base pairs: a density-functional-theory based treatment. J Chem Phys 114:5149–5155
Hill G, Forde G, Hill N, Lester WA Jr, Andrzej Sokalski W, Leszczynski J (2003) Interaction energies in stacked DNA bases? How important are electrostatics? Chem Phys Lett 381:729–732
Jurečka P, Šponer J, Černý J, Hobza P (2006) Benchmark database of accurate (MP2 and CCSD(T) complete basis set limit) interaction energies of small model complexes, DNA base pairs, and amino acid pairs. Phys Chem Chem Phys 8:1985–1993
Šponer J, Jurečka P, Marchan I, Luque FJ, Orozco M, Hobza P (2006) Nature of base stacking: reference quantum-chemical stacking energies in ten unique B-DNA base-pair steps. Chem Eur J 12:2854–2865
Waller MP, Robertazzi A, Platts JA, Hibbs DE, Williams PA (2006) Hybrid density functional theory for π-stacking interactions: application to benzenes, pyridines, and DNA bases. J Comput Chem 27:491–504
Ducéré J-M, Cavallo L (2007) Parametrization of an empirical correction term to density functional theory for an accurate description of π-stacking interactions in nucleic acids. J Phys Chem B 111:13124–13134
Hill JG, Platts JA (2007) Spin-component scaling methods for weak and stacking interactions. J Chem Theor Comput 3:80–85
Langner KM, Sokalski WA, Leszczynski J (2007) Intriguing relations of interaction energy components in stacked nucleic acids. J Chem Phys 127:111102
Swart M, van der Wijst T, Fonseca Guerra C, Bickelhaupt FM (2007) π-π stacking tackled with density functional theory. J Mol Model 13:1245–1257
Wang Y (2008) Theoretical evidence for the stronger ability of thymine to disperse SWCNT than cytosine and adenine: self-stacking of DNA bases vs. their cross-stacking with SWCNT. J Phys Chem C 112:14297–14305
Biczysko M, Panek P, Barone V (2009) Toward spectroscopic studies of biologically relevant systems: vibrational spectrum of adenine as a test case for performances of long-range/dispersion corrected density functionals. Chem Phys Lett 475:105–110
Morgado CA, Jurecka P, Svozil D, Hobza P, Sponer J (2010) Reference MP2/CBS and CCSD(T) quantum-chemical calculations on stacked adenine dimers. Comparison with DFT-D, MP2.5, SCS(MI)-MP2, M06-2X, CBS(SCS-D) and force field descriptions. Phys Chem Chem Phys 12:3522–3534
Rose IA, Hanson KR, Wilkinson KD, Wimmer MJ (1980) A suggestion for naming faces of ring compounds. Proc Natl Acad Sci USA 77:2439–2441
Chen J, Kohler B (2014) Base stacking in adenosine dimers revealed by femtosecond transient absorption spectroscopy. J Am Chem Soc 136:6362–6372
Florián J, Šponer J, Warshel A (1999) Thermodynamic parameters for stacking and hydrogen bonding of nucleic acid bases in aqueous solution: Ab Initio/Langevin Dipoles Study. J Phys Chem B 103:884–892
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06 functionals and 12 other functionals. Theor Chem Acc 120:215–241
Broo A, Holmén A (1996) Ab initio MP2 and DFT calculations of geometry and solution tautomerism of purine and some purine derivatives. Chem Phys 211:147–161
Plutzer C, Kleinermanns K (2002) Tautomers and electronic states of jet-cooled adenine investigated by double resonance spectroscopy. Phys Chem Chem Phys 4:4877–4882
Plutzer C, Nir E, de Vries MS, Kleinermanns K (2001) IR-UV double-resonance spectroscopy of the nucleobase adenine. Phys Chem Chem Phys 3:5466–5469
El Hassan MA, Calladine CR (1997) Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Phil Trans R Soc Lond A 355:43–100
Boys SF, Bernardi F (1970) Calculation of small molecular interactions by differences of separate total energies—some procedures with reduced errors. Mol Phys 19:553–566
Sponer J, Hobza P (1994) Nonplanar geometries of DNA bases. Ab initio Second-Order Moller–Plesset Study. J Phys Chem 98:3161
Peng C, Ayala PY, Schlegel HB, Frisch MJ (1996) Using redundant internal coordinates to optimize equilibrium geometries and transition states. J Comput Chem 17:49–56
Peng C, Schlegel HB (1993) Combining synchronous transit and quasi-Newton methods for finding transition states. Isr J Chem 33:449–454
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision A. 02. Gaussian Inc., Wallingford, CT
Dickerson RE (1989) Definitions and nomenclature of nucleic acid structure components. Nucl Acids Res 17:1797–1803
Widom JR, Johnson NP, von Hippel PH, Marcus AH (2013) Solution conformation of 2-aminopurine dinucleotide determined by ultraviolet two-dimensional fluorescence spectroscopy. New J Phys 15:025028
Smith DA, Holroyd LF, van Mourik T, Jones AC (to be published)
Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE (1981) Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci USA 78:2179–2183
Acknowledgments
We are grateful to EaStCHEM for computational support via the EaStCHEM Research Computing Facility.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
van Mourik, T., Hogan, S.W.L. DNA base stacking involving adenine and 2-aminopurine. Struct Chem 27, 145–158 (2016). https://doi.org/10.1007/s11224-015-0708-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-015-0708-3