Abstract
Purpose
Cover crops and reduced tillage are two key climate smart agricultural practices that can provide agroecosystem services including improved soil health, increased soil carbon sequestration, and reduced fertilizer needs. Crop residue carbon traits (i.e., lignin, holocellulose, non-structural carbohydrates) and nitrogen concentrations largely mediate decomposition rates and amount of plant-available nitrogen accessible to cash crops and determine soil carbon residence time. Non-destructive approaches to quantify these important traits are possible using spectroscopy.
Methods
The objective of this study was to evaluate the efficacy of spectroscopy instruments to quantify crop residue biochemical traits in cover crop agriculture systems using partial least squares regression models and a combination of (1) the band equivalent reflectance (BER) of the PRecursore IperSpettrale della Missione Applicativa (PRISMA) imaging spectroscopy sensor derived from laboratory collected Analytical Spectral Devices (ASD) spectra (n = 296) of 11 cover crop species and three cash crop species, and (2) spaceborne PRISMA imagery that coincided with destructive crop residue collections in the spring of 2022 (n = 65). Spectral range was constrained to 1200 to 2400 nm to reduce the likelihood of confounding relationships in wavelengths sensitive to plant pigments or those related to canopy structure for both analytical approaches.
Results
Models using laboratory BER of PRISMA all demonstrated high accuracies and low errors for estimation of nitrogen and carbon traits (adj. R2 = 0.86 − 0.98; RMSE = 0.24 − 4.25%) and results indicate that a single model may be used for a given trait across all species. Models using spaceborne imaging spectroscopy demonstrated that crop residue carbon traits can be successfully estimated using PRISMA imagery (adj. R2 = 0.65 − 0.75; RMSE = 2.71 − 4.16%). We found moderate relationships between nitrogen concentration and PRISMA imagery (adj. R2 = 0.52; RMSE = 0.25%), which is partly related to the range of nitrogen in these senesced crop residues (0.38–1.85%). PRISMA imagery models were also influenced by atmospheric absorption, variability in surface moisture content, and some presence of green vegetation.
Conclusion
As spaceborne imaging spectroscopy data become more widely available from upcoming missions, crop residue trait estimates could be regularly generated and integrated into decision support tools to calculate decomposition rates and associated nitrogen credits to inform precision field management, as well as to enable measurement, monitoring, reporting, and verification of net carbon benefits from climate smart agricultural practice adoption in an emerging carbon marketplace.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Climate smart agricultural (CSA) practices have emerged as a primary mechanism to improve the sustainability of farming in the face of destabilizing factors linked to a changing climate. Two of the top-tier CSA practices include (1) planting of winter cover crops which can improve soil health and reduce erosion and nutrient loss (Abdalla et al., 2019; Nouri et al., 2022; Wallander et al., 2021), increase soil carbon sequestration (Abdalla et al., 2019; Jian et al., 2020; Zhang et al., 2022) and decrease greenhouse gas emissions (Abdalla et al., 2019; Muhammad et al., 2019); and (2) conservation tillage, which maintains soil structure via a decrease in mechanical tilling of the soil, leaving high levels non-photosynthetic vegetation (NPV) on the soil surface in the form of crop residue thereby reducing soil erosion, preserving or increasing soil carbon, and retaining soil moisture (Du et al., 2022; Follett, 2001; West & Post, 2002). Despite the importance of these two practices, large uncertainties exist in quantifying both the performance of cover crops—quantity (aboveground biomass) and biochemical quality (nitrogen and carbon concentrations)—and the distribution and biochemical quality of crop residues, all of which vary in space and time (Finney et al., 2016; Hively et al., 2009; Pepe et al., 2023; Thapa et al., 2018a, 2018b).
In the past 20 years, multispectral remote sensing has emerged as a viable tool to quantify and map cover crop performance and crop residue cover. Multispectral remote sensing measures reflectance across the visible (V; 400–700 nm), near infrared (NIR; 701–1399 nm), and shortwave infrared (SWIR; 1400–2500 nm) regions of the electromagnetic spectrum in broad, discrete spectral bands (~ 15–180 nm bandwidth). Visible and near infrared (VNIR) spectral indices calculated using multispectral imagery have been used to estimate cover crop aboveground biomass in a variety of regions and species (Bendini et al., 2024; Jennewein et al., 2022; Roth et al., 2023; Thieme et al., 2020; Yuan et al., 2019). However, few studies have sought to quantify cover crop quality—often referred to as “biochemical traits”—using multispectral remote sensing and those that do have focused solely on nitrogen. For instance, recent work demonstrated cover crop nitrogen content may be estimated with moderate to high accuracy (Miller & Shober, 2024; Thieme, 2022; Xia et al., 2021), while estimating nitrogen concentration using multispectral instruments remains challenging (Holzhauser et al., 2022; Roth et al., 2023; Thieme, 2022). These studies leverage the use of the red-edge region (690–790 nm) to estimate nitrogen, but also demonstrate that nitrogen estimates made using multispectral vegetation indices vary between years and seasons (Miller & Shober, 2024; Xu et al., 2018) as well as among cover crop species and geographic region (Xia et al., 2021).
Multispectral remote sensing has also enabled quantification of crop residue fractional cover (Hively et al., 2018; Lamb et al., 2022; Yue et al., 2022), with many studies implementing spectral indices derived from broad SWIR bands from Landsat and Sentinel-2 sensors such as the normalized difference tillage index (NDTI; Van Deventer et al., 1997). While useful for single image analyses, NDTI is subject to interference from moisture content and green vegetation, which greatly influences its utility in multi-temporal analyses (Hively et al., 2019). The novel SWIR angle index may also make improvements on NDTI and can be calculated with the broad spectral bands on Landsat and Sentinel-2 sensors (Yue et al., 2022). Instruments with capacity for narrow-band SWIR measurements can support the calculation of spectral indices that capture lignocellulose absorption features located at 2100 and 2300 nm, enabling highly accurate estimate of residue cover via the shortwave infrared normalized difference residue index (SINDRI) and cellulose absorption index (CAI) (Nagler et al., 2000; Serbin et al., 2009). However, spaceborne instruments with this capacity are limited. Future missions—such as Landsat Next and Sentinel Next Generation—will provide appropriately placed, narrow spectral bands to calculate CAI at scale (Dennison et al., 2023; Hively et al., 2021; Lamb et al., 2022), but these are not yet in operation. The bulk of studies investigating remote sensing approaches for crop residues focus on the quantification of fractional cover (Verrelst et al., 2023). No studies to date focus on quantifying the biochemical quality of crop residues from either cash or cover crops, which has important implications for nutrient cycling and availability for precision management of agroecosystems. A new suite of spaceborne imaging spectroscopy instruments now enables the investigation of crop residue quality and quantity.
The accurate quantification of crop residue biochemical quality offers an opportunity to provide spatially explicit nutrient concentrations that may enable decomposition predictions and inform precision management decisions. For instance, after cover crops are terminated, they can provide part of the nitrogen requirements of the subsequent cash crop thereby offsetting external fertilizer needs (Thapa et al., 2018a, 2018b; Vogeler et al., 2022). A recent multi-state on-farm study showed that cover crop residue carbon traits such as cellulose and hemicellulose (together “holocellulose”), lignin, and non-structural carbohydrates (NSC) are important predictors of residue decomposition rate (along with nitrogen) (Thapa et al., 2018a, 2018b). The presence of corn (Zea mays L.) residue also impacts the timing of available nutrients and is slow to decompose due to its high carbon to nitrogen ratio and largely recalcitrant carbon fractions (Hadas et al., 2004; Wilhelm et al., 2004). Furthermore, residues with higher lignin concentration are less capable of stabilizing soil organic matter (Stewart et al., 2015). In contrast, higher quality residues such as those formed from legume cover crops or multi-species mixes are more likely to be stabilized in mineral-associated organic matter pools (Cotrufo & Lavallee, 2022; Cotrufo et al., 2013; Rocci et al., 2021; Zhang et al., 2022), which increases soil carbon residence time (Cotrufo & Lavallee, 2022; Rocci et al., 2021). Therefore, it is critical to quantify and map the concentrations of these important carbon traits to better understand crop residue decomposition and nutrient release dynamics in cover crop-based cropping systems, which also have important implications for the carbon residence time in the soil (Lavallee et al., 2019; Thapa et al., 2023).
One viable option to accomplish this is the use of spectroscopy (also referred to as “hyperspectral”) instruments, which employ narrow-contiguous spectral collections across the optical spectrum (~ 400–2500 nm). As these spectral bands are contiguous, they provide more detailed spectral information, compared to multispectral instruments, that enable identification of absorption features linked to plant biochemical and biophysical properties (Curran, 1989; Kokaly et al., 2009; Youngentob et al., 2012). Previous work demonstrated that imaging spectroscopy outperformed multispectral imagery in cash crop biomass and nitrogen content estimations in agroecosystems (Lu et al., 2020). Despite such findings, the number of studies employing multispectral imagery far exceed those using imaging spectroscopy due to limited data availability, technical challenges associated with large data volumes, and poor signal-to-noise ratios in heritage imaging spectroscopy systems (Daughtry et al., 2005; Lu et al., 2020). Furthermore, a recent review paper showed only ~ 3.7% of crop nitrogen studies that employed spectroscopy instruments used spaceborne sensors (Fu et al., 2021), thus demonstrating an urgent need to evaluate modern spaceborne imaging spectroscopy systems for nitrogen concentration estimation. Only one study to date has explored the use of spectroscopy instruments to estimate cover crop biomass and nitrogen in cereal rye (Secale cereale L.) while employing airborne imaging spectroscopy (Wang et al., 2023).
Prior studies that have sought to quantify carbon traits in living vegetation have demonstrated mixed success (Asner & Martin, 2015; Chlus & Townsend, 2022; Jennewein et al., 2020; Van Cleemput et al., 2018; Wang et al., 2020). This is likely due to the complex biochemical structure of living vegetation as carbon compounds are intertwined in plant tissues making their separation challenging (Verrelst et al., 2023), particularly in the SWIR region where trait reflectance signatures are obscured by foliar water content (Elvidge, 1990; Kokaly et al., 2009; Thulin et al., 2014). The spectral signatures of photosynthetically active living vegetation compared to NPV are strikingly different, with absorption features of cellulose and lignin more readily discernible as water content decreases (Daughtry et al., 2010; Kokaly et al., 2009). It may be possible to quantify and map these important carbon trait metrics in senesced crop residues following cover crop termination or harvest operations because plant water content is minimal. To our knowledge, no studies have sought to quantify the biochemical quality of crop residues using spectral data, which can greatly improve our understanding of existing nutrients that may be available to the subsequent cash crop in these systems as well as the estimated timing of their release, which is a key component in enhancing precision agricultural practices.
Thus, the overarching aim of this study was to evaluate the efficacy of spectroscopy instruments to quantify crop residue biochemical traits in cover crop agriculture systems in the Mid-Atlantic region of the United States of America (USA). With the recent launch of several spaceborne imaging spectroscopy instruments such as the Italian Space Agency’s Hyperspectral PRecursore IperSpettrale della Missione Applicativa (PRISMA) sensor (Cogliati et al., 2021) and the planned launch of the National Aeronautics and Space Administration’s (NASA) Surface Biology and Geology (SBG) mission and the European Space Agency’s (ESA) Copernicus Hyperspectral Imaging Mission for the Environment (CHIME) missions, there is an urgent need to explore the use of spectroscopy data for estimating crop residue biochemical traits to better enable precision crop and soil management across farmlands. Therefore, the objectives of this study were to: 1) use laboratory-collected spectra (convolved to the band equivalent reflectance of PRISMA) to predict crop residue biochemical traits (i.e., nitrogen, NSC, lignin, and holocellulose) in 11 cover crop species and three cash crop species; and 2) assess the feasibility of spaceborne PRISMA imagery to accurately quantify and map these traits in agricultural fields.
Materials and methods
Study area and field sampling
Data were collected from agricultural fields at the Beltsville Agricultural Research Center (BARC) located in Beltsville Maryland, USA, and on farmers’ fields located on Eastern Shore of Maryland (Table 1; Fig. 1) and are available in a U.S. Geological Survey data release (Hively et al., 2024). Both study locations are in the Mid-Atlantic Coastal Plain, with fine-loamy ultisols (Christiana, Matapeake, and Keyport soil series are predominant at BARC, and Othello and Mattapex series are predominant on the Eastern Shore of Maryland). Cover and cash crops in this region are grown under rainfed conditions. Cover crops are usually planted in October and terminated in March, April, or May and experience an average 530 mm precipitation, and accumulation of 998 growing degree days (GDD) between October 1 and April 30, with 73.5 days below the typical growth threshold for cereals (4 °C). Cash crops including corn and soybean (Glycine max (L.) Merr.) are usually planted in May and harvested in October, and experience 587 mm precipitation and 2843 GDD on average. Small grain cash crops such as winter wheat (Triticum aestivum L.) are planted in September or October and harvested in July and experience 890 mm precipitation and 2647 GDD on average.
Study area in Maryland, USA. Two PRecursore IperSpettrale della Missione Applicativa (PRISMA) images were obtained in April (left; located at the Beltsville Agricultural Research Service) and May (right; located on the Eastern Shore of Maryland) 2022. PRISMA images are shown as true color composites. Field sampling of crop residues (n = 65; red triangles) and nadir photograph acquisition occurred within seven days after PRISMA image acquisition. The right hand PRISMA image is a mosaic of five image tiles, only one of which corresponded to our field sampling locations (Color figure online)
For this study, we collected 296 cover crop and cash crop samples spanning one experimental and two observational trials. For all studies, we destructively sampled each species from a 0.57 m−2 quadrat or a 1-m length of three consecutive rows with a 19-cm row spacing and oven dried the samples to a constant weight at 50–60 °C. Among the three studies, we sampled residue from three cash crop species (corn, wheat, and soybean) as well as 11 cover crop species spanning three common functional groups: grasses, legumes, and brassicas. Although soybean is a type of legume, we chose not to group these samples with the cover crop legumes—crimson clover (Trifolium incarnatum), hairy vetch (Vicia villosa Roth), and winter pea (Pisum sativum)—because we observed significant differences in the concentration of biochemical traits used in this study between leguminous cover crops and soybean samples (Fig. 10, Online Appendix). Table 1 contains a summary of all three data sources and associated spectral collections. Please refer to Table 5 in the Online Appendix for plot and field management details for each trial including seeding rate, planting and termination dates, herbicide, and fertilization details when applicable.
Data from the experimental trial (n = 57) were originally included in Daughtry et al. (2010) and contained wheat residues collected post-harvest on 22 July 2006 at BARC. The purpose of Daughtry et al. (2010) was to quantify changes in fiber concentration in varying stages of decomposition and their associated changes in spectral signatures. Thus, these samples were collected over a two-year period from 2006 to 2008. Approximately half of these samples experienced normal “dry” field conditions (n = 31) while the other half underwent additional wetting to accelerate decomposition (n = 26). The bags of wheat residue were staked securely to the soil surface in a production field on 1 August 2006 in six clusters in a randomized complete block design (two moisture levels and three blocks). The “wet” treatment received 4 mm of irrigation water twice daily for 70 days in 2006 and 91 days in 2007 while the “dry” treatment received only normal precipitation. Air temperatures and precipitation were recorded at a nearby weather station. After 0, 57, 136, 212, 325, and 522 days in the field, six bags of residue were removed from the field (for each date), dried at 50 °C to constant weight, and stored until day 673 when the remaining 18 bags of wheat residues were removed from the field and dried. For the first day of the trial (August 1, 2006) only three samples were included, all from the dry treatment, because the wet treatment also began on this day. Samples were stored in a cool dry location inside of BARC facilities to avoid additional decay between field sampling and spectral collections. Spectral data for this trial included laboratory Analytical Spectral Devices (ASD; Boulder, CO, USA) scans (refer to Sects. “Laboratory spectral collections” for complete details), which were collected for all samples after the conclusion of the trial on day 673. After scanning, these samples were then analyzed for lignin, cellulose, and hemicellulose concentration following the Goering and Van Soest sequential fiber analysis (Van Soest et al., 1991) with modifications by Ankom Technology. No nitrogen or NSC concentrations were quantified in these samples. For the complete details of this study, please refer to Daughtry et al. (2010).
The first observational trial used in this study was comprised of small-plot cover crop plantings (10 × 4 m; 1 sample per plot; n = 112; Table 1) located at BARC that were sampled twice in March of 2022 and twice in April of 2022. This study had a diversity of cover crop species including two grasses—oats (Avena fatua) and cereal rye; three legumes—crimson clover, hairy vetch, and winter pea; and three brassicas—rapeseed (Brassica napus), field mustard/turnip (Brassica rapa), and cabbage/kale (Brassica oleracea) planted in monoculture stands. All samples from this trial were from living cover crops that were destructively sampled as green, photosynthetically active vegetation (i.e., “cut green”), and then oven dried prior to spectral collections. They therefore retained some of their greenness and chlorophyll related features, compared to field-weathered residue. Spectral data collected for this trial included laboratory ASD scans taken immediately after oven drying to a constant weight.
The second observational trial included in this study was a field-scale cover crop plantings of four grasses: cereal rye, wheat, triticale (Triticale hexaploide L.), and barley (Hordeum vulgare), and two cash crop plantings (corn and soybean). Nineteen fields were sampled (eight on the Eastern Shore of Maryland and 11 at BARC) with three to ten locations sampled per field (n = 127; Table 1). This field-scale trial was designed to correspond with spaceborne imaging spectroscopy data; thus, sampling locations were spaced at least 30 m apart and 30 m away from field edges. Approximately half of the samples collected for this field scale trial (n = 62) were living cover crops that were destructively sampled and then oven dried to create residue (i.e., “cut green”). For this subset, we only present the spectral data from laboratory ASD scans in this study because in situ they would not be considered residue (as they would be living green plants in the PRISMA imagery). The remaining half (n = 65) were “true” residues that were either cover crops previously terminated in the field, or crop residue remaining on the fields after harvest operations. These “true” residues senesced in the field and displayed little greenness or evidence of chlorophyll (Fig. 2). They were also oven dried to remove moisture and then scanned in the laboratory with an ASD. Only this subset of “true residue” data is presented in both laboratory-collected ASD scans and spaceborne PRISMA imagery in this study. These samples were collected in the field on May 4th for BARC (n = 36; four fields B1–B4) and on May 26th for the Eastern shore of Maryland (n = 29; eight fields ES1–8).
A representative image from in situ photography from each of the 12 study fields that pair with PRecursore IperSpettrale della Missione Applicativa (PRISMA) imagery. Fields with “ES” label are located on the Eastern Shore of Maryland, while fields with a “B” label are located at the Beltsville Agricultural Research Station (BARC). Numbers in each photograph indicate the calculated fractional green vegetation (GV) cover represented in each image. [image by (W. Dean Hively and Jyoti Jennewein)]
Residue samples from both observational trials (small plot and field scale) were sent to the University of Georgia Feed and Environmental Water Laboratory for near-infrared spectroscopy measurements of crude protein, lignin, cellulose, hemicellulose, and NSC concentrations. In these samples (n = 239), nitrogen concentration was estimated as crude protein divided by 6.25, which is an estimate of the average grams of crude protein required to contain one gram of nitrogen. Holocellulose was estimated by summing cellulose with hemicellulose. Due to the variety of management practices in the three different trials (Table 1) we collected field data from (i.e., small plot, field level, and decomposition), we tested whether the range of observed (measured) lignin and holocellulose varied among them using analysis of variance (ANOVA) and Tukey’s Honest Significant Difference (HSD) post hoc analysis (Tables 6 and 7, Online Appendix). Note, we did not test for inter-trial differences in nitrogen or NSC because the Daughtry et al. (2010) data did not have these measurements included.
In total we collected 296 samples, where 174 were “cut green” living cover crops that were destructively sampled as green photosynthetically active vegetation and then oven dried to create residue, including all samples from the small plot trial and approximately half the samples field scale trial (Table 1). The remaining 122 were “true” in situ residues; either cover crops terminated in the field and left to senesce, or the remains of cash crop harvest operations. Sixty-five of these true residue samples were collected within one week after acquisition of PRISMA images on April 28th and May 21st, 2022 (Fig. 1). These samples were geolocated with either an ArrowGold GPS unit (Eos Positioning System, Inc., Terrebonne QC, Canada) for the BARC samples or a Bad Elf GNSS GPS (Bad Elf GNSS Receivers, Scottsdale, AZ, USA) unit for the Eastern Shore samples.
For the field scale trial data, we collected a series of nadir photographs from each sampled field using either a 12.1-megapixel red–green–blue (RGB) Canon PowerShot G16 camera or a 20.1-megapixel Canon Powershot G7X Mark iii camera (Fig. 2). The camera was mounted on a three-meter painter’s pole. We used these photographs to visualize the conditions of the study fields in reference to the collected PRISMA imagery to calculate fractional green vegetative cover (GV) using a threshold-based approach for red:green ratio (Saberioon et al., 2014) and excess green (Mao et al., 2003) spectral indices.
Laboratory spectral collections
Laboratory spectral collections occurred in a specialized optics laboratory at BARC and followed the protocol detailed in Daughtry et al. (2010). In brief, dried residue samples were placed on a 2 cm depth in 45-cm square trays that were painted a spectrally flat black. Reflectance spectra (350–2500 nm, 2 nm wavelength region at 2 nm intervals, which were interpolated to 1 nm) were acquired using an ASD FieldSpec Pro spectrometer for Daughtry et al. (2010) samples and an ASD FieldSpec-4 Hi-Res spectrometer for all other samples. The fiber optic cable of the spectrometer with attached 18° fore-optic was mounted 60 cm above the collection area with a 0° zenith angle (Fig. 11, 12D and E, Online Appendix). To illuminate the samples, we used six 100 W quartz-halogen lamps mounted 45 cm above the samples with a 45° zenith angle. Output of the lamps was stabilized with a current-regulated DC power supply. For each sample, we collected four spectra (50 integrated scans per spectra) rotating the tray 90° between each spectrum. For each sample, we then calculated the mean of these four spectra to generate a single spectrum. Periodically, a white Spectralon reference panel (Spectralon Labsphere, Inc., North Sutton, NH, USA) was illuminated and viewed in same manner as the samples and was used to convert the collected spectra to percent reflectance.
Laboratory spectral processing
The ASD spectrometers used in this study contain three separate detectors, which sometimes experience small offsets where the wavelengths overlap. The first detector ranged from 350–1000 nm, the second detector from 1001–1800 nm, and the third detector from 1801–2500 nm. Generally, the second detector has been shown to be the most stable of the three (Kokaly & Skidmore, 2015) and therefore the first and third detectors were corrected to better align with the second detector. To achieve this, we replicated correction techniques detailed in Kokaly and Skidmore (2015) in the statistical software R (R Core Team, 2022). To correct the first detector, we conducted a linear interpolation from 1001 and 1002 nm (start of the second detector) to 1000 nm (end of the first detector). Next, we calculated a multiplicative factor by dividing the linearly extrapolated reflectance value by the measured reflectance value at 1000 nm. The multiplicative factor was then applied to all wavelengths of the first detector. We conducted this same process to correct the third detector, but this correction occurred using 1799 and 1800 nm to extrapolate 1801 nm. An example of this correction is available in Fig. 13, Online Appendix.
We calculated the band equivalent reflectance (BER) of PRISMA using the corrected laboratory spectra to assess the potential performance of PRISMA spectral bands. We obtained PRISMA band wavelengths and full-width half maximum (FWHM) values from image metadata and calculated a generalized Gaussian function to project its spectral response function based on these values (Cogliati et al., 2021). We then calculated the weighted mean of the laboratory spectral data and the spectral response of the PRISMA bands using the following equation:
where \({\lambda }_{max}\) and \({\lambda }_{min}\) represent the highest and lowest wavelength of a given PRISMA band, \({r}_{i}\) is the PRISMA spectral response (Gaussian curve based on FWHM) at wavelength \(i\), and \({\rho }_{i}\) denotes the laboratory collected reflectance value at wavelength \(i\). An example of this transformation is supplied in Fig. 14, Online Appendix. Finally, we calculated the first and second derivatives following equations seven and eight, respectively, in Tsai and Philpot (1998) to compare to the corrected and convolved laboratory reflectance spectra (Fig. 12, Online Appendix).
Imagery collection and processing
Two PRISMA images were acquired on April 28th and May 21st, 2022, coinciding with destructive crop residue sampling dates (n = 65) in corn, soybean, cereal rye, barley, wheat, and triticale fields. PRISMA imagery has a reported smile of ~ 2 nm and ~ 5 nm in the VNIR and SWIR respectively (Cogliati et al., 2021). Because our sampling locations were located near the center of the collected PRISMA image tracks, we expect the role of smile to be less extreme than locations near the scene edges, and therefore chose not to apply a smile correction. We downloaded the surface reflectance cubes (L2D) for these images from the PRISMA mission portal and converted them into geotiffs using the R-based ‘prismaread’ package (Busetto & Ranghetti, 2020). We manually adjusted for georectification errors in the PRISMA imagery to obtain a more accurate location on the earth’s surface by applying a singular pair of latitude–longitude corrections to the entire image. We spectrally smoothed the georectification-corrected L2D images to remove noise following Marshall et al. (2022), Tagliabue et al. (2022), and Berger et al. (2021). First, we used the ‘pracma’ package to identify and remove erroneous peaks in each spectrum and then employed the ‘FieldSpectroscopyCC’ package to conduct spectral smoothing using a smoothing spline to all reflectance values using 60 degrees of freedom (Borchers, 2019; Julitta et al., 2016) (Fig. 3). Next, we applied a pixel-wise brightness normalization following the recommendations in Feilhauer et al. (2010). Finally, we removed bands associated with atmospheric constituents with strong absorption features from oxygen, water, carbon dioxide, and methane including 940–962 nm, 1120–1152 nm, 1340–1460 nm, 1785–2019 nm, and 2357–2490 nm.
Example of the acquired (actual) Level 2d (L2D) surface reflectance PRecursore IperSpettrale della Missione Applicativa (PRISMA) spectra (black), smoothed L2D spectra (red), and laboratory-collected band equivalent reflectance (BER) of PRISMA (blue) for two fields. Panel A shows a recently terminated cereal rye field treated with herbicide (field B1), while panel B depicts an older cereal rye residue field with mixed weed species (field B2). (image by [W. Dean Hively and Jyoti Jennewein]) (Color figure online)
Satellite sensors with a moderate spatial resolution, such as PRISMA (30 m), capture mixed pixels where soil, vegetation, and residue all contribute to the spectral reflectance characteristics of a single pixel (e.g., Fig. 3B). To better understand and quantify in-field variability from mixed pixels, we calculated several spectral indices from the two PRISMA images. We first computed the normalized difference vegetation index (NDVI), which can be calibrated to fractional green vegetation cover (FC) using the following equation (Prabhakara et al., 2015):
using the mean of 855–865 nm and 660–679 nm for NIR and red bands, respectively.
To evaluate residue cover we calculated SINDRI, which can be calibrated to fractional residue cover (FR) as follows (Quemada & Daughtry, 2016):
This formation of SINDRI is based on the WorldView-3 SWIR 6 and SWIR 7 bands. For our formulation, we used the mean reflectance of 2183–2222 nm for SWIR 6 and the mean reflectance of 2237–2283 nm for SWIR 7.
We also used the CAI to evaluate fractional residue cover because it is less biased by green vegetation than SINDRI (Dennison et al., 2019; Hively et al., 2021; Lamb et al., 2022). We used the following formula for CAI:
Finally, we calculated a narrow band moisture index calibrated to relative water content (RWC) (Quemada & Daughtry, 2016) using the following equation:
Of these spectral indices and cover estimates, RWC, FR, and FC were already normalized to range from 0–1. To ensure model coefficients and statistics were comparable, we also rescaled CAI to range from 0–1.
Data analysis
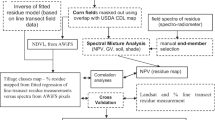
All analyses were conducted using the statistical software program R. For a summary of the processing and analytical steps taken in this work, refer to Fig. 4. We first evaluated whether the mean concentrations of crop residue biochemical traits varied among species functional groups—grasses, legumes, and brassicas for cover crops and corn, grasses, and soybean for cash crops—using ANOVA and Tukey’s HSD post hoc test (Fig. 10, Online Appendix). We also assessed correlations among crop residue traits (Fig. 15, Online Appendix).
Workflow diagram. Colors represent stages of analysis and data inputs with blue denoting data collection, orange denoting laboratory pre-processing and analysis, and green representing true PRecursore IperSpettrale della Missione Applicativa (PRISMA) pre-processing and analysis. Relevant figures, equations, and tables are referenced. ASD Analytical Spectral Devices, L2D spectra Level 2d, PRESS prediction residual sum of squares, RMSE root mean square error, VIP Variable Influence on Projection (Color figure online)
As several dominant absorption features for all residue biochemical traits in this study are located in the SWIR (Curran, 1989; Elvidge, 1990), we built empirical regression models using a 1200–2400 nm range for all traits. Restricting the range of wavelengths also reduces the likelihood of confounding relationships in wavelengths sensitive to plant pigments (400–700 nm) or those related to canopy structure (Asner, 1998; Chlus & Townsend, 2022). For the laboratory collected spectra, we randomly withheld 30% of the samples (n = 70 for nitrogen and NSC; n = 87 for lignin and holocellulose) for model validation. Though selected randomly, the validation dataset spanned a variety of crop species and a wide concentration range for each residue biochemical trait. We further split the remaining 70% of samples into train (70%) and test (30%) splits for calibration using partial least squares regression (PLSR) models in the ‘pls’ package (Wehrens & Mevik, 2007), which were iterated 100 times and had a maximum of 20 components. We took the mean outputs of these 100 iterations to predict the withheld validation data for the BER of PRISMA spectra for all three spectral transformations (i.e., reflectance, first derivatives, and second derivatives) leading to comparison of three models for each of the four-crop residue biochemical traits of interest.
We used the minimum prediction residual sum of squares (PRESS) statistic identified from the calibration data for each validation model to constrain the number of components and minimize the risk of overfitting. To assess the performance of both the calibration and validation models, we used the adjusted coefficient of determination (adj. R2), the root mean square error (RMSE), relative RMSE (%RMSE), Willmott’s index of agreement (d) (Willmott, 1981), and the slope and intercepts of each linear model comparing the observed (measured) value against the predicted value from PLSR models (Piñeiro et al., 2008). Finally, we used the Variable Influence on Projection (VIP; Wold et al., 2001) scores calculated with the ‘plsVarSel’ package (Mehmood et al., 2012) to determine the most important wavelengths for predicting a given residue biochemical trait, which has been used in prior studies of a similar nature (Wang et al., 2021, 2023). To test for functional group differences—corn, soybean, grasses, legumes, and brassicas—we conducted slope and intercept contrast using the ‘emmeans’ package (Lenth et al., 2019).
We also performed a similar set of analyses using spaceborne PRISMA imagery for the sampled locations (n = 65). For these comparisons, we also limited the wavelengths considered to 1200–2400 nm. In addition to individual wavelengths, we included FC, FR, CAI, and RWC spectral indices as predictors in these models to account for variability within a given pixel related to green cover (FC) and residue cover (FR and CAI) and water content (RWC). For these comparisons, we withheld 25% of the samples for validation (n = 17) and iterated train-test splits 100 times. In all comparisons, we continued to evaluate performance using adjusted R2, RMSE, %RMSE, d, and slopes and intercepts of the linear model comparing measured vs. predicted values. Any negative predictions were truncated at zero. We then used model coefficients to produce a calibrated image for the cover crop residue biochemical traits of interest, which was then masked to remove pixels with high green vegetative cover (NDVI > 0.4).
Finally, to compare model coefficients (observed ~ predicted) from actual PRISMA imagery to the laboratory BER of PRISMA, we conducted slope and intercept contrasts using the ‘emmeans’ package. These contrasts enabled us to assess whether model slopes and intercepts varied between laboratory and PRISMA imagery models. To further test whether laboratory simulated (BER PRISMA) values varied from in-field PRISMA imagery, we also subset the laboratory spectra to include only samples corresponding to spaceborne PRISMA imagery. Similar to the above descriptions, we then compared performance using adjusted R2, RMSE, %RMSE, d, and slopes and intercepts and conducted slope and intercept contrasts.
Results
Physical sample concentrations of nitrogen (0.37–4.28%), NSC (6.66–86.34%), and holocellulose (7.93–99.71%) varied significantly between grasses and all other functional groups (legume, brassicas, corn, and soybean; Fig. 10, Online Appendix). No significant differences between legume and brassica traits were observed in this study. Legumes and brassicas had higher nitrogen and NSC concentrations when compared to grasses, corn, and soybean but lower holocellulose concentrations. Lignin concentrations (1.70–26.12%) did not vary statistically between legumes and grasses, but did for grasses and brassicas, corn, and soybeans. We also observed that grasses had a much wider range of lignin concentrations (1.70–26.12%) compared to the brassicas (5.58–9.45%), legumes (5.01–9.45%), corn (10.39–13.92%), and soybean (11.52 –22.21%), likely due to the lignification of grass stems at maturity. We found strong correlations between NSC and holocellulose (r = -0.93), nitrogen and NSC (r = 0.85), and nitrogen and holocellulose (r = -0.82); and weak correlations between holocellulose and lignin (r = 0.01) and nitrogen and lignin (r = -0.22) (Fig. 15, Online Appendix). All reported concentrations were derived from the destructively sampled residues quantified for biochemical traits. No statistically significant differences between trial types (i.e., small plot, field scale, and decomposition) were observed for lignin and holocellulose concentrations (Tables 6 and 7, Online Appendix) indicating that varied management among cash and cover crops had a minimal influence on the biochemical traits in this study.
BER PRISMA Laboratory results
We scanned 296 residue samples from 11 cover crop species and three cash crop species (Table 1) as described above and convolved the collected ASD spectra to the BER of PRISMA (Eq. 1). All crop residue traits showed strong validation results for all three data formats (reflectance and first and second derivatives), with adjusted R2 = 0.863 − 0.978 and %RMSE = 6.306 − 19.730% for BER of PRISMA (Table 2; Fig. 5). Statistical results varied minimally between validation and calibration results (delta adjusted R2 ~ 0.03 for all models), thus we present only validation results. Slopes of all traits were close to one (0.919–1.064) and intercepts ranged from (-0.595–0.930) in all models.
Validation results from partial least squares regression of the band equivalent reflectance PRecursore IperSpettrale della Missione Applicativa (PRISMA) bands to predict the concentration of nitrogen (A–C), nonstructural carbohydrates (NSC) (D–F), holocellulose (G–I), and lignin (J–L) using reflectance (A, D, G, J), first derivative (B, E, H, K), and second derivative (C, F, I, L) in cover crop and cash crop residues. Triangles indicate that the sample is a true, aged residue left in the field to decompose whereas circles indicate samples were collected as living vegetation then dried to create ‘residue.’ RMSE root mean square error (Color figure online)
To assess whether residue biochemical trait models varied between crop functional groups (i.e., grasses, legumes, brassicas, corn, and soybean), we compared linear model coefficients (observed vs. predicted) using slope and intercept contrasts. Because reflectance, first derivative, and second derivative models all demonstrated similar results (Table 2; Fig. 5), we opted to conduct these contrast assessments for the reflectance models only (Fig. 5A, D, G, J). Results showed no statistical differences in slope or intercept among residue functional groups for all residue biochemical traits: nitrogen (t-ratio = − 0.239 − 1.582, p ≥ 0.396), NSC (t-ratio = − 2.124 − 0.698, p ≥ 0.223), holocellulose (t-ratio = − 0.726 − 1.001, p ≥ 0.854), or lignin (t-ratio = − 0.786 − 0.452, p ≥ 0.991).
Wavelengths identified by VIP scores as important (values > 0.8) generally corresponded to known absorption features for each cover crop residue trait (Fig. 6). For instance, nitrogen reflectance models (Fig. 6A, black) showed high VIP scores (> 1.5) near 1500 nm (Elvidge, 1990); while the first and second derivative models showed the highest VIP scores near 1690 and 2300 nm (Curran, 1989; Elvidge, 1990; Youngentob et al., 2012). NSC first and second derivative models also showed high VIP scores near 1690 and 2300 nm (Fig. 6B), likely because NSC is highly correlated with nitrogen in our samples (r = 0.85; Fig. 15, Online Appendix). Similarly, high VIP scores were also observed for lignin and holocellulose at 1690 nm, with holocellulose also demonstrating a high correlation with nitrogen (r = − 0.82; Fig. 15, Online Appendix). This 1690 nm absorption feature is shared by lignin, NSC, and proteins (e.g., nitrogen) (Youngentob et al., 2012). Both nitrogen and NSC also demonstrated high VIP scores from ~ 1170–1300 nm, particularly for reflectance models, but, of the two traits, only NSC has known absorption features in this range near 1200 nm (Curran, 1989). We also observed high VIP scores near known absorption features for cellulose (Fig. 6C) and lignin (Fig. 6D) at 2040, 2100, and 2300 nm. The 2100 and 2300 nm absorption features for cellulose and lignin are widely used and well-documented (Daughtry & Hunt, 2008; Elvidge, 1990; Hively et al., 2021; Lamb et al., 2022; Serbin et al., 2009).
Variable Influence on Projection (VIP) scores for the band equivalent reflectance PRecursore IperSpettrale della Missione Applicativa (PRISMA) bands for nitrogen (A), nonstructural carbohydrates (NSC) (B), holocellulose (C), and lignin (D) in cover crop and cash crop residues. Each of the three data formats is presented on each panel with black representing reflectance, purple representing the first derivative, and green denoting the second derivative. Hashed vertical lines mark known absorption features identified in previous works for each trait (Color figure online)
A subset of our crop residue samples from four grass cover crops and two cash crop species (n = 65; Table 1) were collected within seven days of two PRISMA images. These samples are considered “true” residues that were terminated and left in the field to decompose. We also collected laboratory spectra of these samples, which were convolved to the BER of PRISMA and included in PLSR models report in Table 3 and Fig. 5. With this subset of true residue samples, we also explored whether BER of PRISMA laboratory model statistics and coefficients varied from the complete BER of PRISMA dataset (that also included cut green vegetation dried to create residue). Results for lignin and holocellulose models continued to show strong validation results for the true residue subset of BER of PRISMA, with adjusted R2 = 0.936 and 0.878 and %RMSE = 12.294% and 2.862%, respectively (Table 3). However, we observed a decrease in explained variance for NSC and nitrogen models, with adjusted R2 = 0.693 and 0.780, when the BER of PRISMA models were subset to include only true residues (Table 3). As the relationships for nitrogen and NSC were greatly reduced in this comparison, we expanded the range of wavelengths to include VNIR in a second set of models as it is well documented that nitrogen has additional absorption features in the VNIR, particularly in the red-edge region (~ 680 nm to 730 nm) (Clevers & Gitelson, 2013; Li et al., 2014; Ye et al., 2023). However, the inclusion of VNIR wavelengths did not result significant changes in model predictions or error for nitrogen (adjusted R2 = 0.658, %RMSE = 18.258%) nor NSC (adjusted R2 = 0.704, %RMSE = 17.169%) for this subset of true, aged residue samples with comparatively small range of nitrogen concentration (Fig. 16, Online Appendix).
Results with PRISMA imagery
Using spaceborne PRISMA imagery spectra acquired in the spring of 2022 from two scenes, we assessed biochemical trait predictions for the same subset of true residue data in Table 3. We only present reflectance model results from PRISMA imagery reflectance models because the spectral derivatives demonstrated substantial noise (Fig. 17, Online Appendix). We observed strong relationships between observed vs. predicted NSC, holocellulose, and lignin (adjusted R2 = 0.650, 0.746, 0.748; RMSE = 3.753%, 3.444%, 2.714%; d = 0.884, 0.918, 0.925, respectively; Table 4; Fig. 7). However, nitrogen models performed only moderately well (adjusted R2 = 0.507, RMSE = 0.249%, d = 0.815), potentially due to limitations of the range of values present in these samples (0.376–1.849%). We also explored whether nitrogen or NSC models improved with the addition of VNIR wavelengths, as we did with the subset of BER of PRISMA samples that corresponded to PRISMA imagery (Table 3). With the addition of VNIR to nitrogen and NSC models, we observed modest improvements in accuracy for nitrogen and NSC (adjusted R2 = + 0.02 and + 0.06, respectively) with minimal changes in error, d, or model coefficients (Table 4).
Validation results of PRecursore IperSpettrale della Missione Applicativa (PRISMA) imagery reflectance predicted, using 1200–2400 nm, cover crop residue traits: nitrogen, nonstructural carbohydrates (NSC), holocellulose, and lignin. For each metric, 25% of the field observations (n = 17) were withheld and predicted from the remaining 75% of the data 100 times (mean values of these predictions are displayed). Different colors represent different fields (n = 12). RMSE = root mean square error. Fields with “ES” label are located on the Eastern Shore of Maryland, while fields with a “B” label are located at the Beltsville Agricultural Research Station (BARC) (Color figure online)
Crop residue samples paired with PRISMA imagery (n = 65) had a restricted range for each trait when compared to the full laboratory BER PRISMA dataset (n = 239 − 296; Table 1; Fig. 16, Online Appendix). To assess how linear model coefficients varied from laboratory BER of PRISMA to actual PRISMA imagery, we conducted slope and intercept contrasts. We observed no statistically different slopes or intercepts between the laboratory and in-field models for all cover crop residue nitrogen (T-ratio = − 1.153, p = 0.251), NSC (T-ratio = − 0.089, p = 0.929), holocellulose (T-ratio = 0.231, p = 0.818), or lignin (T-ratio = 0.156, p = 0.876). We also conducted slope and intercept contrasts to compare the BER of PRISMA laboratory data subset for samples that also had corresponding PRISMA imagery. These results also showed no statistical differences in slope or intercepts between models (Figs. 18–21, Online Appendix).
When examining the PRISMA imagery model predictions graphically (Fig. 7), we observed that crop residue biochemical traits varied substantially within some of our sampled fields (e.g., field B3) but not others. The field with the lowest observed variability in all residue biochemical traits (i.e., field B1) also had the most consistent residue cover (87–100% FR derived from SINDRI) (Quemada & Daughtry, 2016) without any influence from weeds (field B2; 3–57% FC from RGB photographs) or regrowing of terminated cover crops (fields B4 and B3; 5–18% FC from RGB photographs). Fields ES4, ES5, and ES7 also displayed variability and were subject to the influence of early growth stage corn (in situ FC = 10 − 19% from RGB photographs). We observed small inter-field variability in lignin concentrations.
We generated calibrated trait maps of our Eastern Shore of Maryland study region (Fig. 8) using SWIR only relationships shown in Fig. 7 and Table 4. Pixels displayed in the map were filtered to include only low NDVI values (< 0.4). This masking ensured minimal influence of vegetation cover in these fields, retaining 59% (B1), 43% (B2), 91% (B3), 68% (B4), 87% (ES1), 93% (ES2), 99% (ES3), 97% (ES4), 94% (ES5), 90% (ES6), 81% (ES7), and 88% (ES8) of pixels for each field scale study field.
Calibrated images of cover crop residue traits: A nitrogen, B nonstructural carbohydrates (NSC), C holocellulose, and D lignin for the Eastern Shore of Maryland fields. The 21 May 2022 PRecursore IperSpettrale della Missione Applicativa (PRISMA) scene was masked to normalized difference vegetation index (NDVI) of < 0.4 to ensure minimal influence of vegetation cover in these fields (Color figure online)
VIP scores of the PRISMA imagery analysis showed that FR was the most important predictor across all residue biochemical traits (Fig. 22, Online Appendix; VIP ≥ 5.90) indicating the amount of fractional residue cover on the satellite derived measurements is highly important for predicting carbon biochemical traits. Other spectral indices—CAI, RWC, and FC—also demonstrated high VIP scores for these traits (VIP > 4.29, VIP > 2.68, VIP > 2.71, respectively). We also observed higher VIP scores near the masked atmospheric absorption regions near 1350 nm and 1450 nm for some traits, which may be related to sensor noise and atmospheric correction errors. VIP scores for the PRISMA imagery analysis were more consistent across the SWIR for all biochemical traits when compared to the laboratory BER results. Because of this, we also visualized standardized mean PLSR coefficients to identify locations that align with previously reported spectral features (Fig. 9).
Standardized partial least squares regression (PLSR) coefficients for cover crop residue nitrogen (A), nonstructural carbohydrates (B), holocellulose (C), and lignin (D) models with PRecursore IperSpettrale della Missione Applicativa (PRISMA) imagery. Hashed vertical lines mark absorption features identified in previous works for each trait
Standardized mean PLSR coefficients showed that previously identified absorption features were present within our crop residue samples (Fig. 9). For instance, we observed an absorption feature near 1690 nm for NSC and lignin that previously has been affiliated with starches and lignin (Curran, 1989). Our results also demonstrate the well-known lignocellulose absorption features located at 2100 nm and 2300 nm (Daughtry & Hunt, 2008; Elvidge, 1990; Hively et al., 2021; Lamb et al., 2022; Nagler et al., 2000; Serbin et al., 2009). These coefficient plots also show that starting at 2000 nm there is an increase in noise, which is also visible when viewing the normalized spectra (Fig. 3).
Discussion
Spectroscopy provides moderate to highly accurate estimations of crop residue biochemical traits, providing important linkages to precision agriculture
In this study, we explored the use of spectroscopy data to predict and map cash and cover crop residue biochemical traits (i.e., nitrogen, NSC, holocellulose, and lignin concentrations) using ASD convolved spectra to simulate PRISMA bands (i.e., BER of PRISMA) as well as using spaceborne PRISMA satellite imagery acquired over two agricultural regions of Maryland in Spring of 2022 (Fig. 1). As expected, BER of PRISMA collected in the laboratory provided highly accurate predictions of all crop residue biochemical traits (Table 2; Fig. 5). Commonly cited absorption features for traits of interest proved to be important in these predictions (Fig. 6) including 1690 nm documented for carbohydrates, and lignin absorption (Curran, 1989), as well as 2100 and 2300 nm for lignin and cellulose (Daughtry & Hunt, 2008; Elvidge, 1990; Hively et al., 2021; Lamb et al., 2022; Nagler et al., 2000; Serbin et al., 2009) and 1200 nm for lignin and cellulose (Curran, 1989). Of note, our results demonstrate that a single empirical model may be used to predict a given residue biochemical trait irrespective of the crop species and functional groups (grasses, legumes, brassicas, corn, and soybeans) as we observed no major differences in the slope or intercepts of the reflectance models between functional groups (Fig. 5; Table 2). Future work could investigate whether including younger grasses, which have greater NSC and lower holocellulose, into models would result in similar findings.
The consistency of PLSR models across functional groups has important practical implications for future efforts in estimating and mapping cover and cash crop residue biochemical traits. For instance, our results suggest a single model can be used to provide input of crop residue biochemical traits, regardless of species, into support tools that aid farmers with decision making regarding residue persistence and the timing of nutrient availability to cash crops. Cover crop functional groups (i.e., grass, legume, brassica) dictate the type of agroecosystem provision, including the rate of residue decomposition and subsequent availability of nutrients to cash crops (Dabney et al., 2001) and the formation and stabilization of soil organic carbon (Zhang et al., 2022). For instance, grass and brassica cover crops play an important role in scavenging residual nitrogen and thereby mitigate nutrient leaching and indirect nitrous oxide emissions, and suppressing weeds (Abdalla et al., 2019; Dabney et al., 2001; Thapa et al., 2018a, 2018b). Winter annual legumes produce nitrogen that is released during decomposition, which in turn can reduce fertilizer requirements (Ranells & Wagger, 1996; Thapa et al., 2022). Because our results show that a single PLSR model may be used across species for a given trait, we hypothesize that we could apply this same approach to multi-species mixtures whose performance and dominant species vary within each planted field as a result of intrinsic (i.e., soil, topography, and weather conditions) and extrinsic (i.e., management decisions) factors (Thapa et al., 2018a, 2018b). This is critical as many cover crop adopters plant multi-species cover crop mixtures as opposed to single species monocultures (Sustainable Agriculture Research and Education (SARE) et al., 2023), due to their ability to provide multiple agroecosystem benefits. Once derived, cover crop trait quantification analysis can be integrated into farm decision support tools such as the existing cover crop nitrogen calculator tool (CC-NCALC; https://covercrop-ncalc.org/). Imaging spectroscopy can help to characterize spatial variability in these traits, driving decision making related to variability in nutrient release relative to soil topographic and textural variability thereby supporting precision nutrient management.
To evaluate the likelihood of these strong laboratory-based relationships enduring in spaceborne imaging spectroscopy collections, we conducted slope and intercept contrasts with the BER of PRISMA reflectance models and the actual PRISMA imagery models’ predictions of cover crop residue biochemical traits. Although the accuracy, errors, and agreements for PRISMA imagery models (Table 4) were not as strong as the relationships observed with the laboratory BER of PRISMA results (Table 2), we found no significant differences between slopes nor intercepts for all crop residue biochemical traits of interest. We further tested this by sub-setting the laboratory samples that coincided with PRISMA imagery (n = 65; Table 3) and found the same result: no significant slope or model intercepts between laboratory-based BER of PRISMA and true PRISMA imagery with this subset of samples. This suggests that spaceborne imaging spectroscopy may be used to map crop residue biochemical traits across a variety of concentrations and species using calibration equations derived in the laboratory, and that the decreased accuracy observed in the imagery analysis was due to complexity of the land surface (influence of exposed soil and green vegetation). Future work could test this hypothesis on a broad range of cash and cover crop species and spaceborne imaging spectroscopy platforms as our in-situ samples contained only grasses, corn, and soybeans although our laboratory models suggest that these relationships would hold across a broader set of species (Fig. 5).
Importantly, PRISMA imagery results for holocellulose and lignin in non-photosynthetic cover crop residues showed substantial improvements (Table 4, Fig. 7) over previous works that quantified these carbon traits in living vegetation (Asner & Martin, 2015; Chlus & Townsend, 2022; Van Cleemput et al., 2018; Wang et al., 2020). Our high accuracy in estimating these carbon traits in non-photosynthetic vegetation is likely due to omission of any influence caused by plant water content, present in living vegetation, that typically masks spectroscopic absorption features in the SWIR region (Elvidge, 1990; Kokaly et al., 2009; Thulin et al., 2014). Previous works that have quantified NSC, cellulose, and lignin in living vegetation using imaging spectroscopy have shown mixed results. For instance, NSC has been accurately estimated across eastern North American biomes (R2 = 0.72; Wang et al., 2020), but in Amazonia NSC accuracy was reduced substantially (R2 = 0.49; Asner & Martin, 2015). Lignin concentration of living vegetation has also proven to be challenging to estimate across regions and vegetation types (R2 = 0.47 − 0.62; Asner & Martin, 2015; Chlus & Townsend, 2022; Jennewein et al., 2020; Van Cleemput et al., 2018; Wang et al., 2020). Our results suggest that applying imaging spectroscopy to senesced, non-photosynthetic cover crop residues (usually present in fields in the springtime following cover crop termination or post-harvest—July for wheat and fall for corn and soybean) provides a unique opportunity to quantify these traits that would otherwise be masked in living vegetation, which in turn can support precision nutrient management (e.g., for example, precision nitrogen application at time of side dress).
Previous works often combined lignin and cellulose together into a single “lignocellulose” or NPV category because they are intimately intertwined within plant tissues and therefore many of their spectral absorption features overlap (Kokaly et al., 2009). These characterizations are useful for estimating residue cover (Daughtry et al., 2004, 2005; Pepe et al., 2020, 2023), which has important implications for carbon cycling and erosion control. However, these broad categorizations are not amenable to distinguishing recalcitrant carbon fractions such as lignin from labile, readily decomposable sources of carbon from NSC. Though carbon concentration has been shown to be relatively consistent among plant litters, the partitioning of carbon to specific compounds such as lignin, cellulose, and NSC is critical for accurate predictions of residue decomposition and subsequent nitrogen release rates (Cotrufo et al., 2013; Daughtry et al., 2010; Thapa et al., 2022). Furthermore, crop residue biochemical traits influence their residence time in the soil (Lavallee et al., 2019; Thapa et al., 2023). Higher quality residues (low carbon to nitrogen (C:N) ratio and higher NSC) are more likely to be stabilized in mineral-associated organic matter pools, whereas lower quality plant residues primarily form particulate organic matter (Cotrufo & Lavallee, 2022; Cotrufo et al., 2013; Rocci et al., 2021; Zhang et al., 2022). Mineral-associated organic matter is a more persistent form of organic matter capable of retaining greater amounts of soil carbon (Cotrufo & Lavallee, 2022; Rocci et al., 2021).
Cover crops and conservation tillage practices provide two powerful CSA practices that may increase soil carbon sequestration, but their biomass and biochemical composition exert great influence on this process (Jian et al., 2020). Therefore, an accurate quantification of the concentration of each carbon trait is useful to characterize the resulting type of soil organic carbon formed, soil and water health, and the timing of nutrient release to the cash crop (Cotrufo et al., 2022; Dabney et al., 2001). As routine acquisitions of spaceborne imaging spectroscopy become widely available towards the end of the decade through platforms like NASA’s SBG and the ESA’s CHIME satellites, we anticipate that robust calibrations may be generated to predict these important biochemical traits in cover crop residues. When these capabilities are integrated into decision support tools, it can facilitate precision management of mineral fertilizer inputs (Thapa et al., 2022) as well as enable measurement, monitoring, reporting, and verification of net carbon and greenhouse gas benefits from CSA practice adoption in an emerging carbon marketplace.
In contrast to our results for carbon-based residue traits, our estimates of in-field nitrogen concentrations in true residues were only moderately successful (adj. R2 = 0.507, %RMSE = 19.857%; d = 0.815), compared to results for the full range of fresh-dried and field-aged samples (adj. R2 = 0.905, %RMSE = 11.802%; d = 0.975). Numerous studies have shown accurate estimates of nitrogen concentration in photosynthetically-active living vegetation (e.g., Van Cleemput et al., 2018; Wang et al., 2023) even while using only SWIR wavelengths (Chlus & Townsend, 2022). In fact, a recent study demonstrated that nitrogen concentration can be accurately estimates across a wide variety and crops and regions using imaging spectroscopy (adj. R2 = 0.78; Dai et al., 2023). One likely explanation for our results is the limited range of nitrogen concentration within the field-collected true residue samples (0.376–2.053%; Fig. 16, Online Appendix), which were all grass, soybean, and corn residue that tend to have lower nitrogen concentrations when compared to brassica and legume species (Fig. 10, Online Appendix). This explanation is supported by our laboratory results, where one can observe poorer model performance when laboratory data are subset for only true residues (Table 3). This strongly suggests that given a wider range of values our predictions could be improved. Future work could include a diverse range of species timed with spaceborne imaging spectroscopy to broaden the sampled range of this important trait. Even with a limited range of nitrogen concentration in our true residue samples, our PLSR results are similar to those found using multispectral remote sensing (Holzhauser et al., 2022; Xia et al., 2021).
Another plausible explanation for this finding may be that these are “true” aged cover crop residues—they were terminated and left in the field to decompose—and therefore plant chlorophyll had fully degraded, changing spectral features of nitrogen and protein. Our results with VNIR and SWIR wavelengths support this, as we observed a modest increase in accuracy when VNIR wavelengths were incorporated with SWIR wavelengths although error, agreement, and model coefficients were quite similar (Table 4). This is likely because in true decomposing residues, spectral features present in living vegetation are substantially altered. For instance, chlorophyll absorption is largely absent in the red-edge region (~ 680 nm to 730 nm) sensitive to nitrogen (Clevers & Gitelson, 2013; Li et al., 2014; Wang et al., 2023) (Fig. 12A, Online Appendix). This would suggest that nitrogen concentration may be more easily estimated in living cover crops rather than in residues.
Future work addressing variability related to mixed pixels and moisture content could help clarify complex agroecosystem characteristics
Mixed pixels and moisture content variability in PRISMA imagery likely impacted our results. Although our results demonstrate that cash and cover crop residue carbon traits can be accurately estimated using spaceborne imaging spectroscopy (Table 4), we observed substantial variation in field conditions among sampled locations (Fig. 2). Four of our study fields had weeds or early growth stage corn growing while two had more visible soil background; both of which influenced the spectral signatures of sampled residue locations. To understand the effects of these mixed pixels, we calculated fractional vegetative and residue cover using existing and well-studied spectral indices (Prabhakara et al., 2015; Quemada & Daughtry, 2016). VIP scores for PRISMA imagery models (Fig. 22, Online Appendix) revealed that FR was the most important predictor for crop residue carbon trait predictions, followed by the CAI, RWC, and FC. Work that addresses spatial and temporal heterogeneity of mixed pixels could help improve predictions of crop residue traits in future investigations.
Spectral mixture analysis is one viable method to resolve issues related to mixed pixels (Adams et al., 1986; Keshava & Mustard, 2002). Spectral mixture analysis has been widely employed to unmix spectral collections into their respective sources—or “endmembers”—which are typically composed of soil, residue, vegetation, and shadow (Somers et al., 2011). However, recent work suggests that empirical models (such as PLSR) may outperform spectral mixture analysis techniques in FR cover estimation (Dennison et al., 2019). In this study, we achieved better results with laboratory spectroscopy (Table 2; Fig. 5) than for PRISMA imagery (Table 4, Fig. 7) for all crop residue biochemical traits in part due to limited range of biochemical concentrations for this subset of data (Table 3; Fig. 16, Online Appendix), but also due to mixing of ground cover components (i.e., residue, green vegetation, soil) as evidenced by high VIP score for spectral index proxies for ground cover (FR, CAI, FC; Fig. 22, Online Appendix). Future work could first perform spectral mixture analysis to determine the composition of each pixel, then apply statistical models to predict cover crop residue traits on the unmixed residue component; or alternatively predict FR using PLSR. Furthermore, the characterization of in-scene residue moisture content is critical because moisture strongly affects residue decomposition rates (Thapa et al., 2021) and can also influence the depth of important spectral absorption features (Kokaly et al., 2009; Pepe et al., 2023). Future work could develop moisture correction techniques to address these limitations.
Future work to further correct L2D PRISMA reflectance data could improve data quality
Although PRISMA spectra have a fairly high signal-to-noise ratios (VNIR > 160; SWIR > 100), “spikiness” is evident in the L2D spectra, and many previous works applied spectral smoothing as a pre-processing technique to L2D imagery (Berger et al., 2021; Marshall et al., 2022; Tagliabue et al., 2022). We also followed these recommendations in this study for L2D scenes. Cogliati et al. (2021) conducted the first evaluation of the spectral, spatial, and radiometric performance of PRISMA top of atmosphere radiance (L1) and demonstrated that L1 products are of high quality; however, they suggest L2D product quality also needs further assessment. In this study, we observed substantial noise in the L2D PRISMA spectra prior to smoothing (Fig. 3; Fig. 17, Online Appendix). Many of the peaks identified in the smoothing process were located near known atmospheric absorption features including: 940–962, 1120–1152, 1340–1460, and 1785–2019 nm (Fig. 23, Online Appendix). This suggests that further work to correct for the influence of atmospheric constituents in L2D PRISMA products could improve data quality and may explain some of the observed differences between laboratory simulated (BER PRISMA) and PRISMA imagery models observed in this study.
One possible avenue to correct this is by leveraging recently established NASA spectroscopy tools. One such tool is the Space-based Imaging Spectroscopy and Thermal pathfindER (SISTER) (Townsend et al., 2020). In the SISTER processing pipeline, L1 radiance imaging spectroscopy data undergo a complex series of radiometric and geometric corrections to account for spectral non-uniformities from rotations or shifts in the instrument, smile, and global positioning error. After these steps, corrected radiance images can be atmospherically corrected using the Imaging Spectrometer optimal FITting (ISOFIT) (Thompson et al., 2018) codebase, which employs a radiative transfer model emulator for MODerate resolution atmospheric TRANsmission (MODTRAN) (Berk et al., 2014). Since both SISTER and ISOFIT codebases are freely available on GitHub, future work could apply and compare these techniques to currently used methods for PRISMA imagery processing. Alternatively, other atmospheric correction techniques including Fast Line-of-sight Atmospheric Analysis of Hypercubes (FLAASH) or MODerate resolution atmospheric TRANsmission (MODTRAN) could be evaluated to assess which method would generate the best correction method.
Conclusions and implications
Quantifying and mapping cash crop and cover crop residue biochemical traits is highly important to estimate the magnitude of agroecosystems services that crop residues provide including the amount of carbon stored and stabilized in different soil fractions as well as the provisioning of nutrients to subsequent cash crops following decomposition. This study provided an important step toward the accurate quantification of crop residue biochemical traits using spectroscopy of non-photosynthetic residues, which, apart from nitrogen, have proven to be challenging in photosynthetically active living vegetation due to confounding influence from plant water content. Our work demonstrates that accurate estimates of crop residue carbon-based biochemical traits are achievable with spaceborne imaging spectroscopy; a resource that will be widely available towards the end of the decade or in early 2030s via SBG and the CHIME satellites. Such enhanced capabilities can better inform decision support tools that enable precision management of fertilizer inputs and provide enhanced capabilities in measuring, monitoring, reporting, and verification of net carbon benefits from CSA practice adoption in emerging climate-smart marketplaces. Accurate spaceborne predictions of crop residue biochemical traits will help advance the precision and sustainability of agriculture in a changing climate.
Data Availability
Data used in this paper are available in a U.S. Geological Survey data release (Hively et al., 2024).
References
Abdalla, M., Hastings, A., Cheng, K., Yue, Q., Chadwick, D., Espenberg, M., Truu, J., Rees, R. M., & Smith, P. (2019). A critical review of the impacts of cover crops on nitrogen leaching, net greenhouse gas balance and crop productivity. Global Change Biology, 25(8), 2530–2543. https://doi.org/10.1111/gcb.14644
Adams, J. B., Smith, M. O., & Johnson, P. E. (1986). Spectral mixture modeling: A new analysis of rock and soil types at the Viking Lander 1 site. Journal of Geophysical Research, 91(B8), 8098–8112. https://doi.org/10.1029/jb091ib10p10513
Asner, G. P. (1998). Biophysical and biochemical sources of variability in canopy reflectance. Remote Sensing of Environment, 64(3), 234–253. https://doi.org/10.1016/S0034-4257(98)00014-5
Asner, G. P., & Martin, R. E. (2015). Spectroscopic remote sensing of non-structural carbohydrates in forest canopies. Remote Sensing, 7(4), 3526–3547. https://doi.org/10.3390/rs70403526
Bendini, N., Fieuzal, R., Carrere, P., Clenet, H., Galvani, A., Allies, A., & Ceschia, É. (2024). Estimating winter cover crop biomass in france using optical sentinel-2 dense image time series and machine learning. Remote Sensing, 16(834), 1–24. https://doi.org/10.3390/rs16050834
Berger, K., Hank, T., Halabuk, A., Rivera-Caicedo, J. P., Wocher, M., Mojses, M., Gerhátová, K., Tagliabue, G., Dolz, M. M., Venteo, A. B. P., & Verrelst, J. (2021). Assessing non-photosynthetic cropland biomass from spaceborne hyperspectral imagery. Remote Sensing, 13(22), 1–20. https://doi.org/10.3390/rs13224711
Berk, A., Conforti, P., Kennett, R., Perkins, T., Hawes, F., & Van Den Bosch, J. (2014). MODTRAN®6: A major upgrade of the MODTRAN®radiative transfer code. 2014 6th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), https://doi.org/10.1109/WHISPERS.2014.8077573
Borchers, H. W. (2019). pracma: Practical numerical math functions. R Package Version, 2(9), 519.
Busetto, L., Ranghetti, L. (2020). prismaread: A tool for facilitating access and analysis of PRISMA L1/L2 hyperspectral imagery v1.0.0. Retrieved March 11, 2024, from https://irea-cnr-mi.github.io/prismaread/
Chlus, A., & Townsend, P. A. (2022). Characterizing seasonal variation in foliar biochemistry with airborne imaging spectroscopy. Remote Sensing of Environment, 275(March), 113023. https://doi.org/10.1016/j.rse.2022.113023
Clevers, J. G. P. W., & Gitelson, A. A. (2013). Remote estimation of crop and grass chlorophyll and nitrogen content using red-edge bands on sentinel-2 and-3. International Journal of Applied Earth Observation and Geoinformation, 23(1), 344–351. https://doi.org/10.1016/j.jag.2012.10.008
Cogliati, S., Sarti, F., Chiarantini, L., Cosi, M., Lorusso, R., Lopinto, E., Miglietta, F., Genesio, L., Guanter, L., Damm, A., Pérez-López, S., Scheffler, D., Tagliabue, G., Panigada, C., Rascher, U., Dowling, T. P. F., Giardino, C., & Colombo, R. (2021). Remote sensing of environment the PRISMA imaging spectroscopy mission: Overview and first performance analysis. Remote Sensing of Environment. https://doi.org/10.1016/j.rse.2021.112499
Cotrufo, M. F., Haddix, M. L., Kroeger, M. E., & Stewart, C. E. (2022). The role of plant input physical-chemical properties, and microbial and soil chemical diversity on the formation of particulate and mineral-associated organic matter. Soil Biology and Biochemistry. https://doi.org/10.1016/j.soilbio.2022.108648
Cotrufo, M. F., & Lavallee, J. M. (2022). Soil organic matter formation, persistence, and functioning: A synthesis of current understanding to inform its conservation and regeneration. In D. Sparks (Ed.), Advances in agronomy (1st ed., Vol. 172, pp. 1–66). Academic Press.
Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K., & Paul, E. (2013). The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Global Change Biology, 19(4), 988–995. https://doi.org/10.1111/gcb.12113
Curran, P. J. (1989). Remote sensing of foliar chemistry. Remote Sensing of Environment, 30(3), 271–278. https://doi.org/10.1016/0034-4257(89)90069-2
Dabney, S. M., Delgado, J. A., & Reeves, D. W. (2001). Using winter cover crops to improve soil and water quality. Communications in Soil Science and Plant Analysis, 32(7–8), 1221–1250. https://doi.org/10.1081/CSS-100104110
Dai, J., Jamalinia, E., Vaughn, N. R., Martin, R. E., König, M., Hondula, K. L., Calhoun, J., Heckler, J., & Asner, G. P. (2023). A general methodology for the quantification of crop canopy nitrogen across diverse species using airborne imaging spectroscopy. Remote Sensing of Environment. https://doi.org/10.1016/j.rse.2023.113836
Daughtry, C. S. T., & Hunt, E. R. J. (2008). Mitigating the effects of soil and residue water contents on remotely sensed estimates of crop residue cover. Remote Sensing of Environment, 112(4), 1647–1657. https://doi.org/10.1016/j.rse.2007.08.006
Daughtry, C. S. T., Hunt, E. R., Doraiswamy, P. C., & McMurtrey, J. E. (2005). Remote sensing the spatial distribution of crop residues. Agronomy Journal, 97(3), 864–871. https://doi.org/10.2134/agronj2003.0291
Daughtry, C. S. T., Hunt, E. R., & McMurtrey, J. E. (2004). Assessing crop residue cover using shortwave infrared reflectance. Remote Sensing of Environment, 90(1), 126–134. https://doi.org/10.1016/j.rse.2003.10.023
Daughtry, C. S. T., Serbin, G., Iii, J. B. R., Doraiswamy, P. C., & Raymond, E. H., Jr. (2010). Spectral reflectance of wheat residue during decomposition and remotely sensed estimates of residue cover. Remote Sensing, 2(2), 416–431. https://doi.org/10.3390/rs2020416
Dennison, P. E., Lamb, B. T., Campbell, M. J., Kokaly, R. F., Hively, W. D., Vermote, E., & Wu, Z. (2023). Modeling global indices for estimating non-photosynthetic vegetation cover. Remote Sensing of Environment, 295, 113715. https://doi.org/10.1016/j.rse.2023.113715
Dennison, P. E., Qi, Y., Meerdink, S. K., Kokaly, R. F., Thompson, D. R., Daughtry, C. S. T., Quemada, M., Roberts, D. A., Gader, P. D., Wetherley, E. B., Numata, I., & Roth, K. L. (2019). Comparison of methods for modeling fractional cover using simulated satellite hyperspectral imager spectra. Remote Sensing. https://doi.org/10.3390/rs11182072
Du, X., Jian, J., Du, C., & Stewart, R. D. (2022). Conservation management decreases surface runoff and soil erosion. International Soil and Water Conservation Research, 10(2), 188–196. https://doi.org/10.1016/j.iswcr.2021.08.001
Elvidge, C. D. (1990). Visible and near infrared reflectance characteristics of dry plant materials. International Journal of Remote Sensing, 11(10), 1775–1795. https://doi.org/10.1080/01431169008955129
Feilhauer, H., Asner, G. P., Martin, R. E., & Schmidtlein, S. (2010). Brightness-normalized partial least squares regression for hyperspectral data. Journal of Quantitative Spectroscopy and Radiative Transfer, 111(12–13), 1947–1957. https://doi.org/10.1016/j.jqsrt.2010.03.007
Finney, D. M., White, C. M., & Kaye, J. P. (2016). Biomass production and carbon/nitrogen ratio influence ecosystem services from cover crop mixtures. Agronomy Journal, 108(1), 39–52. https://doi.org/10.2134/agronj15.0182
Follett, R. F. (2001). Soil management concepts and carbon sequestration in cropland soils. Soil and Tillage Research, 61(1–2), 77–92. https://doi.org/10.1016/S0167-1987(01)00180-5
Fu, Y., Yang, G., Pu, R., Li, Z., Li, H., Xu, X., Song, X., Yang, X., & Zhao, C. (2021). An overview of crop nitrogen status assessment using hyperspectral remote sensing: Current status and perspectives. European Journal of Agronomy, 124(October 2019), 126241. https://doi.org/10.1016/j.eja.2021.126241
Hadas, A., Kautsky, L., Goek, M., & Kara, E. (2004). Rates of decomposition of plant residues and available nitrogen in soil, related to residue composition through simulation of carbon and nitrogen turnover. Soil Biology and Biochemistry, 36, 255–266. https://doi.org/10.1016/j.soilbio.2003.09.012
Hively, W. D., Jennewein, J., & Lamb, B. T. (2024). Row crop and cover crop residue spectra from lab spectrometer and spaceborne PRISMA imagery. U.S. Geological Survey data release. https://doi.org/10.5066/P13L34JU
Hively, W. D., Lamb, B. T., Daughtry, C. S. T., Serbin, G., Dennison, P., Kokaly, R. F., Wu, Z., & Masek, J. G. (2021). Evaluation of SWIR crop residue bands for the landsat next mission. Remote Sensing, 13(18), 3718. https://doi.org/10.3390/rs13183718
Hively, W. D., Lamb, B. T., Daughtry, C. S. T., Shermeyer, J., McCarty, G. W., & Quemada, M. (2018). Mapping crop residue and tillage intensity using worldview-3 satellite shortwave infrared residue indices. Remote Sensing. https://doi.org/10.3390/rs10101657
Hively, W. D., Lang, M., McCarty, G. W., Keppler, J., Sadeghi, A., & McConnell, L. L. (2009). Using satellite remote sensing to estimate winter cover crop nutrient uptake efficiency. Journal of Soil and Water Conservation, 64(5), 303–313. https://doi.org/10.2489/jswc.64.5.303
Hively, W. D., Shermeyer, J., Lamb, B. T., Daughtry, C. T., Quemada, M., & Keppler, J. (2019). Mapping crop residue by combining landsat and worldview-3 satellite imagery. Remote Sensing, 11(16), 1–21. https://doi.org/10.3390/rs11161857
Holzhauser, K., Räbiger, T., Rose, T., Kage, H., & Kühling, I. (2022). Estimation of biomass and n uptake in different winter cover crops from UAV-based multispectral canopy reflectance data. Remote Sensing, 14(18), 4525. https://doi.org/10.3390/rs14184525
Jennewein, J. S., Eitel, J. U. H., Pinto, J. R., & Vierling, L. A. (2020). Toward mapping dietary fibers in northern ecosystems using hyperspectral and multispectral data. Remote Sensing, 12(16), 2579. https://doi.org/10.3390/rs12162579
Jennewein, J. S., Lamb, B. T., Hively, W. D., Thieme, A., Thapa, R., Goldsmith, A., & Mirsky, S. B. (2022). Integration of satellite-based optical and synthetic aperture radar imagery to estimate winter cover crop performance in cereal grasses. Remote Sensing, 14(9), 2077. https://doi.org/10.3390/rs14092077
Jian, J., Du, X., Reiter, M. S., & Stewart, R. D. (2020). A meta-analysis of global cropland soil carbon changes due to cover cropping. Soil Biology and Biochemistry, 143(January), 107735. https://doi.org/10.1016/j.soilbio.2020.107735
Julitta, T., Migliavacca, M., & Wutzler, T. (2016). FieldSpectroscopyCC: R package for Characterization and Calibration of spectrometers. Retrieved March 11, 2024, from https://github.com/tommasojulitta/FieldSpectroscopyCC
Keshava, N., & Mustard, J. F. (2002). Spectral unmixing. IEEE Signal Processing Magazine, 19(1), 44–57. https://doi.org/10.1109/79.974727
Kokaly, R. F., Asner, G. P., Ollinger, S. V., Martin, M. E., & Wessman, C. A. (2009). Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sensing of Environment, 113(SUPPL. 1), S78–S91. https://doi.org/10.1016/j.rse.2008.10.018
Kokaly, R. F., & Skidmore, A. K. (2015). Plant phenolics and absorption features in vegetation reflectance spectra near 1.66 μm. International Journal of Applied Earth Observation and Geoinformation, 43, 55–83. https://doi.org/10.1016/j.jag.2015.01.010
Lamb, B. T., Dennison, P. E., Hively, W. D., Kokaly, R. F., Serbin, G., Wu, Z., Dabney, P. W., Masek, J. G., Campbell, M., & Daughtry, C. S. T. (2022). Optimizing landsat next shortwave infrared bands for crop residue characterization. Remote Sensing, 14(23), 6128. https://doi.org/10.3390/rs14236128
Lavallee, J. M., Soong, J. L., & Cotrufo, M. F. (2019). Conceptualizing soil organic matter into particulate and mineral-associated forms to address global change in the 21st century. Global Change Biology, 26(1), 261–273. https://doi.org/10.1111/gcb.14859
Lenth, R., Singmann, H., Love, J., Buerkner, P., & Herve, M. (2019). Package ‘emmeans.’ Retrieved March 11, 2024, from https://cran.r-project.org/web/packages/emmeans/index.html
Li, F., Miao, Y., Feng, G., Yuan, F., Yue, S., Gao, X., Liu, Y., Liu, B., Ustin, S. L., & Chen, X. (2014). Improving estimation of summer maize nitrogen status with red edge-based spectral vegetation indices. Field Crops Research, 157, 111–123. https://doi.org/10.1016/j.fcr.2013.12.018
Lu, B., Dao, P. D., Liu, J., He, Y., & Shang, J. (2020). Recent advances of hyperspectral imaging technology and applications in agriculture. Remote Sensing, 12(16), 2659. https://doi.org/10.3390/RS12162659
Mao, W., Wang, Y., & Wang, Y. (2003). Real-time detection of between-row weeds using machine vision. 2003 ASAE Annual International Meeting, Las Vegas, NV. American Society of Agricultural and Biological Engineers. https://elibrary.asabe.org/abstract.asp?aid=15381
Marshall, M., Belgiu, M., Boschetti, M., Pepe, M., Stein, A., & Nelson, A. (2022). Field-level crop yield estimation with PRISMA and Sentinel-2. ISPRS Journal of Photogrammetry and Remote Sensing, 187(February), 191–210. https://doi.org/10.1016/j.isprsjprs.2022.03.008
Mehmood, T., Liland, K. H., Snipen, L., & Sæbø, S. (2012). A review of variable selection methods in partial least squares regression. Chemometrics and Intelligent Laboratory Systems, 118, 62–69. https://doi.org/10.1016/j.chemolab.2012.07.010
Miller, J. O., & Shober, A. L. (2024). Assessing relationships of cover crop biomass and nitrogen content to multispectral imagery. Agronomy Journal. https://doi.org/10.1002/agj2.21554
Muhammad, I., Sainju, U. M., Zhao, F., Khan, A., Ghimire, R., Fu, X., & Wang, J. (2019). Regulation of soil CO2 and N2O emissions by cover crops: A meta-analysis. Soil and Tillage Research, 192(January), 103–112. https://doi.org/10.1016/j.still.2019.04.020
Nagler, P. L., Daughtry, C. S. T., & Goward, S. N. (2000). Plant litter and soil reflectance. Remote Sensing of Environment, 71(2), 207–215. https://doi.org/10.1016/S0034-4257(99)00082-6
Nouri, A., Lukas, S., Singh, S., Singh, S., & Machado, S. (2022). When do cover crops reduce nitrate leaching? A Global Meta-Analysis. Global Change Biology, 28(15), 4736–4749. https://doi.org/10.1111/gcb.16269
Pepe, M., Pompilio, L., Gioli, B., Busetto, L., & Boschetti, M. (2020). Detection and classification of non-photosynthetic vegetation from PRISMA hyperspectral data in croplands. Remote Sensing, 12(23), 3903. https://doi.org/10.3390/rs12233903
Pepe, M., Pompilio, L., Ranghetti, L., Nutini, F., & Boschetti, M. (2023). Mapping spatial distribution of crop residues using PRISMA satellite imaging spectroscopy. European Journal of Remote Sensing, 56(1), 2122872. https://doi.org/10.1080/22797254.2022.2122872
Piñeiro, G., Perelman, S., Guerschman, J. P., & Paruelo, J. M. (2008). How to evaluate models: Observed vs. predicted or predicted vs. observed? Ecological Modelling, 216(3–4), 316–322. https://doi.org/10.1016/j.ecolmodel.2008.05.006
Prabhakara, K., Hively, W. D., & McCarty, G. W. (2015). International Journal of Applied Earth Observation and Geoinformation Evaluating the relationship between biomass, percent groundcover and remote sensing indices across six winter cover crop fields in Maryland, United States. International Journal of Applied Earth Observations and Geoinformation, 39, 88–102. https://doi.org/10.1016/j.jag.2015.03.002
Quemada, M., & Daughtry, C. S. T. (2016). Spectral indices to improve crop residue cover estimation under varying moisture conditions. Remote Sensing. https://doi.org/10.3390/rs8080660
R Core Team. (2022). R: A Language and Environment for Statistical Computing. https://www.r-project.org/
Ranells, N. N., & Wagger, M. G. (1996). Nitrogen release from grass and legume cover crop monocultures and bicultures. Agronomy Journal, 88(5), 777–782. https://doi.org/10.2134/agronj1996.00021962008800050015x
Rocci, K. S., Lavallee, J. M., Stewart, C. E., & Cotrufo, M. F. (2021). Soil organic carbon response to global environmental change depends on its distribution between mineral-associated and particulate organic matter: A meta-analysis. Science of the Total Environment, 793, 148569. https://doi.org/10.1016/j.scitotenv.2021.148569
Roth, R. T., Chen, K., Scott, J. R., Jung, J., Yang, Y., Camberato, J. J., & Armstrong, S. D. (2023). Prediction of cereal rye cover crop biomass and nutrient accumulation using multi-temporal unmanned aerial vehicle based visible-spectrum vegetation indices. Remote Sensing, 15(3), 580. https://doi.org/10.3390/rs15030580
Saberioon, M. M., Amin, M. S. M., Anuar, A. R., Gholizadeh, A., Wayayok, A., & Khairunniza-Bejo, S. (2014). Assessment of rice leaf chlorophyll content using visible bands at different growth stages at both the leaf and canopy scale. International Journal of Applied Earth Observation and Geoinformation, 32(1), 35–45. https://doi.org/10.1016/j.jag.2014.03.018
Serbin, G., Daughtry, C. S. T., Hunt, E. R., Brown, D. J., & McCarty, G. W. (2009). Effect of soil spectral properties on remote sensing of crop residue cover. Soil Science Society of America Journal, 73(5), 1545–1558. https://doi.org/10.2136/sssaj2008.0311
Somers, B., Asner, G. P., Tits, L., & Coppin, P. (2011). Endmember variability in spectral mixture analysis : A review. Remote Sensing of Environment, 115(7), 1603–1616. https://doi.org/10.1016/j.rse.2011.03.003
Stewart, C. E., Moturi, P., Follett, R. F., & Halvorson, A. D. (2015). Lignin biochemistry and soil N determine crop residue decomposition and soil priming. Biogeochemistry, 124, 335–351. https://doi.org/10.1007/s10533-015-0101-8
Sustainable Agriculture Research and Education (SARE), Conservation Technology Information Center (CTIC), and American Seed Trade Association (ASTA). (2023). 2022–2023 National Cover Crop Survey Report. https://www.sare.org/wp-content/uploads/2022-2023-National-Cover-Crop-Survey-Report.pdf
Tagliabue, G., Boschetti, M., Bramati, G., Candiani, G., Colombo, R., Nutini, F., Pompilio, L., Rivera-caicedo, J. P., Rossi, M., Rossini, M., Verrelst, J., & Panigada, C. (2022). Hybrid retrieval of crop traits from multi-temporal PRISMA hyperspectral imagery. ISPRS Journal of Photogrammetry and Remote Sensing, 187(February), 362–377. https://doi.org/10.1016/j.isprsjprs.2022.03.014
Thapa, R., Cabrera, M., Reberg-Horton, C., Dann, C., Balkcom, K. S., Fleisher, D., Gaskin, J., Hitchcock, R., Poncet, A., Schomberg, H. H., Timlin, D., & Mirsky, S. B. (2022). Modeling surface residue decomposition and N release using the cover crop nitrogen calculator (CC - NCALC). Nutrient Cycling in Agroecosystems, 124, 81–99. https://doi.org/10.1007/s10705-022-10223-3
Thapa, R., Cabrera, M., Schomberg, H. H., Reberg-Horton, C., Poffenbarger, H., & Mirsky, S. B. (2023). Chemical differences in cover crop residue quality are maintained through litter decay. PLoS ONE, 18(7), 1–20. https://doi.org/10.1371/journal.pone.0289352
Thapa, R., Mirsky, S. B., & Tully, K. L. (2018a). Cover crops reduce nitrate leaching in agroecosystems: A global meta-analysis. Journal of Environmental Quality, 47(6), 1400–1411. https://doi.org/10.2134/jeq2018.03.0107
Thapa, R., Poffenbarger, H., Tully, K. L., Ackroyd, V. J., Kramer, M., & Mirsky, S. B. (2018b). Biomass production and nitrogen accumulation by hairy vetch–cereal rye mixtures: A meta-analysis. Agronomy Journal, 110(4), 1197–1208. https://doi.org/10.2134/agronj2017.09.0544
Thapa, R., Tully, K. L., Cabrera, M., Dann, C., Schomberg, H. H., Timlin, D., Gaskin, J., Reberg-horton, C., & Mirsky, S. B. (2021). Agricultural and Forest Meteorology Cover crop residue moisture content controls diurnal variations in surface residue decomposition. Agricultural and Forest Meteorology, 308–309(March), 108537. https://doi.org/10.1016/j.agrformet.2021.108537
Thapa, R., Tully, K. L., Reberg-Horton, C., Cabrera, M., Davis, B. W., Fleisher, D., Gaskin, J., Hitchcock, R., Poncet, A., Schomberg, H. H., Seehaver, S. A., Timlin, D., & Mirsky, S. B. (2022). Cover crop residue decomposition in no-till cropping systems: Insights from multi-state on-farm litter bag studies. Agriculture, Ecosystems and Environment, 326, 107823. https://doi.org/10.1016/j.agee.2021.107823
Thieme, A. (2022). Multispectral satellite remote sensing approaches for estimating cover crop performance in Maryland and Delaware [Unpublished manuscript]. Department of Geographical Sciences, University of of Maryland.
Thieme, A., Yadav, S., Oddo, P. C., Fitz, J. M., McCartney, S., King, L. A., Keppler, J., McCarty, G. W., & Hively, W. D. (2020). Using NASA Earth observations and Google Earth Engine to map winter cover crop conservation performance in the Chesapeake Bay watershed. Remote Sensing of Environment, 248(June), 111943. https://doi.org/10.1016/j.rse.2020.111943
Thompson, D. R., Natraj, V., Green, R. O., Helmlinger, M. C., Gao, B. C., & Eastwood, M. L. (2018). Optimal estimation for imaging spectrometer atmospheric correction. Remote Sensing of Environment, 216(July), 355–373. https://doi.org/10.1016/j.rse.2018.07.003
Thulin, S., Hill, M. J., Held, A., Jones, S., & Woodgate, P. (2014). Predicting levels of crude protein, digestibility, lignin and cellulose in temperate pastures using hyperspectral image data. American Journal of Plant Sciences, 5(7), 997–1019. https://doi.org/10.4236/ajps.2014.57113
Townsend, P. A., Gierach, M. M., Brosnan, I. G., Poulter, B., & Schimel, D. (2020). SISTER: SBG Space-based Imaging Spectroscopy and Thermal PathfindER. AGU Fall Meeting Abstracts, 2020, IN041–03. https://ui.adsabs.harvard.edu/abs/2020AGUFMIN041..03T/abstract
Tsai, F., & Philpot, W. (1998). Derivative analysis of hyperspectral data. Remote Sensing of Environment, 66(1), 41–51. https://doi.org/10.1016/S0034-4257(98)00032-7
Van Cleemput, E., Vanierschot, L., Fernández-Castilla, B., Honnay, O., & Somers, B. (2018). The functional characterization of grass- and shrubland ecosystems using hyperspectral remote sensing: Trends, accuracy and moderating variables. Remote Sensing of Environment, 209(January), 747–763. https://doi.org/10.1016/j.rse.2018.02.030
Van Deventer, A. P., Ward, A. D., Gowda, P. M., & Lyon, J. G. (1997). Using thematic mapper data to identify contrasting soil plains and tillage practices. Photogrammetric Engineering and Remote Sensing, 63(1), 87–93.
Van Soest, P. J., Robertson, J. B., & Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. Journal of Dairy Science, 74(10), 3583–3597. https://doi.org/10.3168/jds.S0022-0302(91)78551-2
Verrelst, J., Halabuk, A., Atzberger, C., Hank, T., Steinhauser, S., & Berger, K. (2023). A comprehensive survey on quantifying non-photosynthetic vegetation cover and biomass from imaging spectroscopy. Ecological Indicators, 155(May), 110911. https://doi.org/10.1016/j.ecolind.2023.110911
Vogeler, I., Böldt, M., & Taube, F. (2022). Mineralisation of catch crop residues and N transfer to the subsequent crop. Science of the Total Environment, 810, 152142. https://doi.org/10.1016/j.scitotenv.2021.152142
Wallander, S., Smith, D., Bowman, M., & Claassen, R. (2021). Cover crop trends, programs, and practices in the United States, EIB 222, U.S. Department of Agriculture, Economic Research Service.
Wang, S., Guan, K., Wang, Z., Ainsworth, E. A., Zheng, T., Townsend, P. A., Li, K., Moller, C., Wu, G., & Jiang, C. (2021). Unique contributions of chlorophyll and nitrogen to predict crop photosynthetic capacity from leaf spectroscopy. Journal of Experimental Botany, 72(2), 341–354. https://doi.org/10.1093/jxb/eraa432
Wang, S., Guan, K., Zhang, C., Jiang, C., Zhou, Q., Li, K., Qin, Z., Ainsworth, E. A., He, J., Wu, J., Schaefer, D., Gentry, L. E., Margenot, A. J., & Herzberger, L. (2023). Airborne hyperspectral imaging of cover crops through radiative transfer process-guided machine learning. Remote Sensing of Environment, 285(November 2022), 113386. https://doi.org/10.1016/j.rse.2022.113386
Wang, Z., Chlus, A., Geygan, R., Ye, Z., Zheng, T., Singh, A., Couture, J. J., Cavender-Bares, J., Kruger, E. L., & Townsend, P. A. (2020). Foliar functional traits from imaging spectroscopy across biomes in eastern North America. New Phytologist, 228(2), 494–511. https://doi.org/10.1111/nph.16711
Wehrens, R., & Mevik, B.-H. (2007). The pls package: principal component and partial least squares regression in R. Retrieved March 11, 2024, from https://CRAN.R-project.org/package=pls
West, T. O., & Post, W. M. (2002). Soil organic carbon sequestration rates by tillage and crop rotation. Soil Science Society of America Journal, 66(6), 1930–1946. https://doi.org/10.2136/sssaj2002.1930
Wilhelm, W. W., Johnson, J. M. F., Hatfield, J. L., Voorhees, W. B., & Linden, D. R. (2004). Crop and soil productivity response to corn residue removal: A literature review. Agronomy, 96(1), 1–17. https://doi.org/10.2134/agronj2004.1000a
Willmott, C. J. (1981). On the validation of models. Physical Geography, 2(2), 184–194. https://doi.org/10.1080/02723646.1981.10642213
Wold, S., Sjöström, M., & Eriksson, L. (2001). PLS-regression: A basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems, 58(2), 109–130. https://doi.org/10.1016/S0169-7439(01)00155-1
Xia, Y., Guan, K., Copenhaver, K., & Wander, M. (2021). Estimating cover crop biomass nitrogen credits with Sentinel-2 imagery and sites covariates. Agronomy Journal, 113(2), 1084–1101. https://doi.org/10.1002/agj2.20525
Xu, M., Lacey, C. G., & Armstrong, S. D. (2018). The feasibility of satellite remote sensing and spatial interpolation to estimate cover crop biomass and nitrogen uptake in a small watershed. Journal of Soil and Water Conservation, 73(6), 682–692. https://doi.org/10.2489/jswc.73.6.682
Ye, L., Guan, K., Qin, Z., Wang, S., Zhou, W., Peng, B., Grant, R., Tang, J., Hu, T., Jin, Z., & Schaefer, D. (2023). Improved quantification of cover crop biomass and ecosystem services through remote sensing- based model—data fusion. Environmental Research Letters, 18(9), 094018. https://doi.org/10.1088/1748-9326/ace4df
Youngentob, K. N., Renzullo, L. J., Held, A. A., Jia, X., Lindenmayer, D. B., & Foley, W. J. (2012). Using imaging spectroscopy to estimate integrated measures of foliage nutritional quality. Methods in Ecology and Evolution, 3(2), 416–426. https://doi.org/10.1111/j.2041-210X.2011.00149.x
Yuan, M., Burjel, J. C., Isermann, J., Goeser, N. J., & Pittelkow, C. M. (2019). Unmanned aerial vehicle—based assessment of cover crop biomass and nitrogen uptake variability. Journal of Soil and Water Conservation. https://doi.org/10.2489/jswc.74.4.350
Yue, J., Fu, Y., Guo, W., Feng, H., & Qiao, H. (2022). Estimating fractional coverage of crop, crop residue, and bare soil using shortwave infrared angle index and Sentinel-2 MSI. International Journal of Remote Sensing, 43(4), 1253–1273. https://doi.org/10.1080/01431161.2022.2032454
Zhang, Z., Kaye, J. P., Bradley, B. A., Amsili, J. P., & Suseela, V. (2022). Cover crop functional types differentially alter the content and composition of soil organic carbon in particulate and mineral-associated fractions. Global Change Biology, 28(19), 5831–5848. https://doi.org/10.1111/gcb.16296
Acknowledgements
The authors would like to thank Jordan Nicolette, Caitlin Dubik, Mary LaChina, Saeka Foreman, Avi Goldsmith, Amara Hayling, and Eric Deutsch for their contributions in field and spectral data collection and processing. This research was a contribution from the Long-Term Agroecosystem Research (LTAR) network. LTAR is supported by the U.S. Department of Agriculture. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Funding
This project was supported by the U.S. Geological Survey, Land Change Science Program within the Core Science Systems Mission Area; the U.S. Department of Agriculture—Agricultural Research Service, National Program 211; and the U.S. Department of Agriculture Lower Chesapeake Long Term Agricultural Research (LTAR) program. Additionally, this work is supported by the Agriculture and Food Research Initiative’s Sustainable Agricultural Systems Coordinated Agricultural Project [award no. 2019-68012-29818] and Water Coordinated Agricultural Project [award no. 2018-68011-28372] from the U.S. Department of Agriculture (USDA) National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jennewein, J.S., Hively, W., Lamb, B.T. et al. Spaceborne imaging spectroscopy enables carbon trait estimation in cover crop and cash crop residues. Precision Agric (2024). https://doi.org/10.1007/s11119-024-10159-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s11119-024-10159-4