Abstract
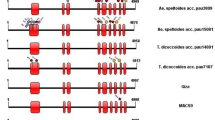
Gelatinization temperature (GT), an important parameter for rice cooking quality, is mainly regulated by the ALK gene encoding starch synthase IIa. Here, we reported the nucleotide diversity of the ALK gene in 122 cultivated accessions and 199 wild rice accessions that were collected around the Pearl River Basin in China. A total of 93 single nucleotide ploymorphisms (SNPs) were identified, with an average of one SNP per 40 bp. Tajima D statistics revealed that the DNA sequences covering the last exon have probably evolved under balancing selection. Based on two functional SNPs (an A to G substitution at 4198 bp and a GC to TT dinucleotide substitution at 4330/4331 bp), three haplotypes, G/GC, G/TT, and A/GC, were identified in both wild and cultivated accessions, with the G/GC haplotype being predominant. Interestingly, the A/GC haplotype was exclusively found in the wild accessions from Guangdong province, while the G/TT haplotype was only present in the wild accessions from Jiangxi province and Hainan Island. This suggests that the G/TT and A/GC variants may have arisen independently and undergone balancing selection on separate haplotypes in multiple populations. Our result supports earlier hypothesis that cultivated rice was independently domesticated from multiple domestication events in China. Our study aids in the understanding of the domestication process that led to the improvement of rice grain quality.


Similar content being viewed by others
References
Abeysekera WKSM, Somasiri HPPS, Premakumara GAS, Bentota AP, Rajapakse D, Ediriweera N (2008) Cooking and eating quality traits of some Sri Lankan traditional rice varieties across Yala and Maha seasons. Trop Agric Res 20:168–176
Bao JS, Sun M, Zhu LH, Corke H (2004) Analysis of quantitative trait locus for some starch properties in rice (Oryza sativa L.), thermal properties, gel texture, swelling volume. J Cereal Sci 39:379–385
Bao JS, Corke H, Sun M (2006) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor Appl Genet 113:1171–1183
Breathnach R, Chambon P (1981) Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Chen LJ, Lee DS, Song ZP, Suh HS, Lu BR (2004) Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann Bot 93:67–73
Cheng J, Khan MA, Qiu WM, Li J, Zhou H, Zhang Q, Guo W, Zhu T, Peng J, Sun F, Li S, Korban SS, Han Y (2012) Diversification of genes encoding granule-bound starch synthase in monocots and dicots is marked by multiple genome-wide duplication events. PLoS One 7:e30088
Ebadi AA, Farshadfar E, Rabiei B (2013) Mapping QTLs controlling cooking and eating quality indicators of Iranian rice using RILs across three years. Aust J Crop Sci 7:1494–1502
Fan CC, Yu XQ, Xing YZ, Xu CG, Luo LJ, Zhang QF (2005) The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor Appl Genet 110:1445–1452
Gao Z, Zeng D, Cui X, Zhou Y, Yan M, Huang D, Li J, Qian Q (2003) Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China C Life Sci 46:661–668
Gao Z, Zeng D, Cheng F, Tian Z, Guo L, Su Y, Yan M, Jiang H, Dong G, Huang Y, Han B, Li J, Qian Q (2011) ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J Integr Plant Biol 53:756–765
Han YP, Xu ML, Liu XY, Yan CJ, Korban SS, Chen XL, Gu MH (2004) Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci 166:357–364
Han Y, Bendik E, Sun FJ, Gasic K, Korban SS (2007a) Genomic isolation of genes encoding starch branching enzyme II (SBEII) in apple: toward characterization of evolutionary disparity in SbeII genes between monocots and eudicots. Planta 226:1265–1276
Han Y, Gasic K, Sun F, Xu M, Korban SS (2007b) A gene encoding starch branching enzyme I (SBEI) in apple (Malus x domestica, Rosaceae) and its phylogenetic relationship to Sbe genes from other angiosperms. Mol Phylogenet Evol 43:852–863
Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, Fan D, Lu Y, Weng Q, Liu K, Zhou T, Jing Y, Si L, Dong G, Huang T, Lu T, Feng Q, Qian Q, Li J, Han B (2011) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44:32–39
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Jiang L, Liu L (2006) New evidence for origins of sedentism and rice domestication in the lower Yangzi River, China. Antiquity 80:355–361
Kumar I, Khush GS (1986) Gene dosage effects of amylose content in rice endosperm. Jpn J Genet 61:559–568
Lanceras JC, Hun ZL, Naivikul Q, Vanavichit A, Ruanjaichon V, Tragoonrung S (2000) Mapping of genes for cooking and eating qualities in Thai Jasmine rice (KDML105). DNA Research 7:93–101
Liu Q, Wang Z, Chen X, Cai X, Tang S, Yu H, Zhang J, Hong M, Gu M (2003) Stable inheritance of the antisense Waxy gene in transgenic rice with reduced amylose level and improved quality. Transgenic Res 12:71–82
Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci 103:9578–9583
Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, Zhang D, Chen C (2015) Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet 128:359–371
Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano HY, Suzuki Y, Sano Y (2008) Allelic diversification at the wx locus in landraces of Asian rice. Theor Appl Genet 116:979–989
Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34:173–185
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleilc Acids Res 8:4321–4325
Nakamura Y, Francisco PB, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N (2005) Essential amino acid of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58:213–227
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Normile D (1997) Archaeology: Yangtze seen as earliest rice site. Science 275:309–310
Panlasigui LN, Thompson LU, Juliano BO, Perez CM, Yiu SH, Greenberg GR (1991) Rice varieties with similar amylose content differ in starch digestibility and glycemic response in humans. Am J Clin Nutr 54:871–877
Phing Lau WC, Latif MA, Y Rafii M, Ismail MR, Puteh A (2014) Advances to improve the eating and cooking qualities of rice by marker-assisted breeding. Crit Rev Biotechnol 17:1–12
Shobha Rani N, Pandey MK, Prasad GSV, Sudharshan I (2006) Historical significance, grain quality features and precision breeding for improvement of export quality basmati varieties in India. Indian J Crop Sci 1:29–41
Song Z, Lu BR, Zhu Y, Chen J (2003) Gene flow from cultivated rice to the wild species Oryza rufipogon under experimental field conditions. New Phytol 157:657–665
SwoVord DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tan YF, Li JX, Yu SB, Xing YZ, Xu CG, Zhang QF (1999) The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor Appl Genet 99:642–648
Thompson J, Chenna Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25:4876–4882
Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D, Wang Y, Yu J, Gu M, Li J (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA 106:21760–21765
Tran DS, Tran TTH, Nguyen TLH, Ha ML, Dinh BY, Kumamaru T, Satoh H (2013) Variation on starch properties and the relationship to single nucleotide polymorphism in SSIIa in waxy rice collected from central of Vietnam. J Fac Agr Kyushu Univ 58:247–252
Umemoto T, Aoki N (2005) Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Functional Plant Biology 32:763–768
Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104:1–8
Umemoto T, Aoki N, Lin HX, Nakamura Y, Inouchi N, Sato Y, Yano M, Hirabayashi H, Maruyama S (2004) Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct Plant Biol 31:671–684
Waters DL, Henry RJ, Reinke RF, Fitzgerald MA (2006) Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol J 4:115–122
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popu Biol 7:256–276
Wei X, Qiao WH, Chen YT, Wang RS, Cao LR, Zhang WX, Yuan NN, Li ZC, Zeng HL, Yang QW (2012) Domestication and geographic origin of Oryza sativa in China: insights from multilocus analysis of nucleotide variation of O. sativa and O. rufipogon. Mol Ecol 21:5073–5087
Yan CJ, Tian ZX, Fang YW, Yang YC, Li J, Zeng SY, Gu SL, Xu CW, Tang SZ, Gu MH (2011) Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.). Theor Appl Genet 122:63–76
Yu G, Olsen KM, Schaal BA (2011) Association between nonsynonymous mutations of starch synthase IIa and starch quality in rice (Oryza sativa). New Phytol 189:593–601
Zeng X (1990) The origin and early development of rice varieties of Jiangxi. Agricultural archaeology 1:166–171
Zong Y, Chen Z, Innes JB, Chen C, Wang Z, Wang H (2007) Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449:459–462
Acknowledgments
This research was jointly supported by the CAS Strategic Priority Research Program (grant no. XDA05130403), and Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (grant no. SAJC201327).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(XLSX 37 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Zheng, H., Wei, G. et al. Nucleotide Diversity and Molecular Evolution of the ALK Gene in Cultivated Rice and its Wild Relatives. Plant Mol Biol Rep 34, 923–930 (2016). https://doi.org/10.1007/s11105-016-0975-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-0975-1