Abstract
Background and Aims
Nitrogen (N) is an essential macronutrient that can limit plant development and crop yield through widespread physiological and molecular impacts. In maize, N-starvation enhances biosynthesis and exudation of strigolactones (SLs) in a process reversible by nitrate addition and consequent repression of genes for SL biosynthesis.
Methods
In the present study, a maize mutant deficient in SL biosynthesis (zmccd8) allowed an in-depth analysis of SL contributions under low N. Both hydroponic and field conditions were used to better characterize the response of the mutant to N availability.
Results
The severity of responses to N-limitation by the SL-deficient zmccd8 mutant extended from growth parameters to content of iron, sulfur, protein, and photosynthetic pigments, as well as pronounced impacts on expression of key genes, which could be crucial molecular target for the SL-mediated acclimatation to N shortage.
Conclusions
Our results demonstrate that SLs are critical for physiological acclimation to N deficiency by maize and identify central players in this action. Further contributions by iron and sulfur are implicated in the complex pathway underlying SL modulation of responses to N-deprivation, thus widening our knowledge on SL functioning and providing new hints on their potential use in agriculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N) is one of the most important macronutrients and limiting factors for plant development, and photosynthetic efficiency (Crawford & Forde 2002; Gojon 2017; Anas et al. 2020). N-shortage in maize reduces both the photochemical capacity of photosystem II (PSII) and the activity of carbon fixation enzymes such as the phosphoenolpyruvate carboxylase (PEPCase), leading to the loss of chlorophyll and soluble protein typical of leaf senescence (Ding et al. 2005; Mu et al. 2017). In wheat, it diminishes the capacity for electron transport from PSII and suppresses tiller outgrowth (Sigalas et al. 2023). On the other hand, excessive N inhibits the synthesis of storage carbohydrates and decreases the content of chlorophyll and soluble proteins (Dechorgnat et al. 2018; Sun et al. 2020). A correct balance of N levels is therefore important to optimize the N use efficiency (NUE), that is crucial for new sustainable agricultural models (Bathaei & Štreimikienė, 2023).
Regardless of the nitrogen form distributed with fertilization, nitrate (NO3−) and ammonium (NH4+) are the two main N forms absorbed by plants (Wani et al. 2021) and NO3− is the preferred source in aerobic agricultural soils (Miller et al. 2007). NO3− also acts as a signal able to affect many developmental and physiological processes (Bouguyon et al. 2012; O'Brien et al. 2016; Maghiaoui et al. 2020). Extensive evidences support the hormonal control of plant responses to NO3−, especially highlighting the auxin stimulation of lateral root development in response to NO3−availability (Yu et al. 2015; Asim et al. 2020; Abualia et al. 2022).
In addition to auxin, strigolactones (SLs) also contribute to the regulation of plant responses to N (Manoli et al. 2016; Marzec & Melzer 2018; Yoneyama 2019). In maize, Ravazzolo et al. (2019, 2021) found that N-deprivation increased SL production and exudation, similarly to what observed in sorghum (Yoneyama et al. 2007), rice (Sun et al. 2014), and Arabidopsis (Ito et al. 2016).
Strigolactones are terpenoid lactones derived from the carotenoid biosynthetic pathway and trigger seed germination for many parasitic plants in the Orobanchaceae family (Cook et al. 1966). SLs also stimulate hyphal branching of arbuscular mycorrhizal fungi (AMF) (Akiyama et al. 2005) and act directly as phytohormones regulating shoot and root architectures (Gomez-Roldan et al. 2008; Umehara et al. 2008). Biosynthesis of SLs is significantly promoted by nutritional deficiency, as for example by the lack of N, phosphorus (P), or sulphur (S) (Yoneyama et al. 2012; Shindo et al. 2018) and SL-mediated root development aids acclimation to nutrient starvation in soils (Ito et al. 2016). SLs are involved in the response to N deficiency in different species (Marzec and Melzer 2018). For instance, in rice, thanks to a crosstalk with gibberellins, SLs positively affect the accumulation of the GROWTH-REGULATING FACTOR 4 (GRF4), a transcription factor that promotes and integrates N assimilation, C fixation and growth (Li et al. 2018), thus leading to improve NUE (Sun et al. 2023). Similarly, in tomato under N-limiting conditions, SLs control the expression of genes involved in NO3−uptake and signalling, such as Nitrate Transporter 1.1 and Nitrate-Inducible (Marro et al. 2022). On the contrary, in many species NO3− provision to N-deprived plants inhibits SL biosynthesis through repressing the expression of different SL biosynthesis genes (D27, CCD7, CCD8, CYP711As) (Barbier et al. 2023).
Guan et al. (2012) identified a maize mutant unable to synthesize SLs (zmccd8), due to the insertion of a Dissociation (Ds) transposon in the third exon of the ZmCCD8 gene that is required for biosynthesis of SLs. zmccd8 plants are shorter with small ears, narrow stalks, a mild branching phenotype, and a smaller root system (Guan et al. 2012). A RNA-seq systematic analyses of the transcriptomic profiles characterizing this mutant (Li et al. 2023) allowed to identify many transcripts encoding proteins with putative roles as SL downstream effectors. Some of these encode components of the transport systems for sulfur (S) and iron (Fe). This is not surprising since the involvement of SLs in the response to S was already hypothesized, as described above (Shindo et al. 2018). Regarding Fe and SL relationship, in hydroponically grown rice plants SL levels were not increased upon Fe starvation, contrary to what observed under P, N and S deficiency (Shindo et al. 2018). However, Song and co-workers (2023) have recently demonstrated that OsNLP4 (NIN-like protein 4), which is crucial to maintain the N-Fe balance, also suppresses SL signalling, providing novel hints on the existence of an interplay among N, Fe and SLs in rice. Furthermore, Fe is an essential component of the first enzymes of SL biosynthesis, namely D27 (Lin et al. 2009), in addition to its involvement in N assimilation enzymes and Fe–S clusters in photosynthetic electron transport (Briat et al. 2015).
In the present study, the zmccd8 mutant was used to better assess the role played by SLs in the maize response to nitrogen. To this aim, seedlings of both genotypes were grown in hydroponics with different NO3−concentration and their growth, and the tissues content of photosynthetic pigments, protein, S and Fe were determined. The two genotypes were also grown in field to observe their phenotype in later development stages. Furthermore, to better understand the molecular basis underlying differences in responses, expression of selected genes was monitored and compared throughout experiments. Genes were selected basing both on previous transcriptomic dataset results (Ravazzolo et al. 2021 and Li et al. 2023) and on their putative functions in the SL biosynthesis/signaling pathways and in processes involved in the transport and assimilation of N, S and Fe. Our results provide new knowledge on the SL-mediated molecular regulation of maize acclimation to nitrogen fluctuations and highlight a previously unrecognized association with changes in content and distribution of sulfur and iron.
Materials and methods
Maize growth conditions in hydroponic conditions and in field
In this study, the maize inbred line B73 (Zea mays L.) and the zmccd8::Ds insertion mutant line in B73 background were utilized. The zmccd8::Ds allele in B73 was created by backcrossing into B73 for 6 generations (Guan et al. 2012). Inbred line B73 (Zea mays L.), and zmccd8 mutant, from now referred as wild-type (WT) and zmccd8 respectively, were germinated for hydroponic as described by Manoli et al. (2014). After germination for four days in paper rolls wet with demineralized water, seedlings were selected to have the highest phenotypic homogeneity and then grown for two (T2) to ten days (T10) in a modified Hoagland nutrient solution (Quaggiotti et al. 2003) with three different concentrations of NO3−: a N-free treatment (0), a KNO3 0.1 mM (0.1) and KNO3 1 mM (1) (Table 1). The duration of the treatments in nutrient solution was chosen based on previous research conducted on the same species which highlighted a clear response to the availability of exogenous NO3− by the roots already at these stages of development and regardless of the nutritional content of the seed (Ravazzolo et al. 2021, 2019; Trevisan et al. 2011, 2012, 2019; Manoli et al. 2014, 2016).
About 15 seedlings were grown in a glass container for each replicate and they were placed in growth chambers with a day/night cycle of 14/10 h at 25/18° C air temperature, 70/90% relative humidity, and 280 μmol \({m}^{-2}{s}^{-1}\) photon flux density (Quaggiotti et al. 2003). Different analyses were performed after two (T2), three (T3), four (T4), six (T6), seven (T7) and ten (T10) days in the nutrient solutions (Table 1). The nutrient solutions were constantly aerated and changed every two days. All sampling or measurements were performed at 11 a.m. after 4 h of light. For each condition, three biological replicates were analysed (15 plants for each condition). All chemicals were purchased from Sigma Chemicals (Sigma, St Louis, MO, USA) unless otherwise stated.
WT and zmccd8 were also grown in open field during the spring–summer 2022 in Azienda Agraria Sperimentale L. Toniolo (Legnaro, PD, ITA). The two genotypes were sown in April and, about 50 days after sowing, the plants were fertilized with urea (N 46%).
Dry weight and length of the shoots and primary roots under hydroponic conditions and phenotypic analysis of maize plant in open field
Roots and leaves of each treatment (0, 0.1, 1 mM NO3−, 15 plants each) were separately sampled at T7 in hydroponics, in three independent biological repetitions, and weighted as both fresh weight and dry weight after seven days at 60 °C. At T7 the leaf development reached the third leaf. Root and leaves images were collected using a flatbed scanner. The length of the shoot and the primary root (PR) was measured by means of ImageJ software analysis (https://imagej.nih.gov). Data represent the average of four independent biological replicates, each replicate considering 6 plants for every treatment (n = 6) ± standard error.
The WT and zmccd8 mutant circumference and height of the stem, the length of the internode and leaves, and the number of leaves were assessed at 46, 56 and 66 days after sowing (das) in the open field. Data represent the average of 24 independent biological replicates, each replicate considers a single plant as a biological replicate for each treatment (n = 24) ± standard error.
For statistical analysis, data were considered significant when p < 0.05. by ANOVA test performed with the Fisher's least significant difference (LSD) multiple comparison method.
Optics measurements of chlorophyll and anthocyanins in the leaf
DUALEX SCIENTIFIC + ™ (Force-A, Orsay, France) was used to evaluate chlorophyll (CHL), anthocyanins (ANT) and the Nitrogen Balance Index (NBI) in both hydroponics conditions and 46–56-66 days after sowing for open field conditions. Two readings were made for each seedling at 2/3 of the distance from the leaf base as suggested by Yuan et al. (2016) on the first leaf after three (T3) and four (T4) days and on the first and second leaf after six (T6), seven (T7) and ten (T10) days in every treatment (0, 0.1, 1 mM NO3−, 15 plants each) for hydroponic conditions. In the open field, two readings on the third leaf for each plant were achieved. Four biological replicates for each treatment and an ANOVA statistic test (p < 0.05) were performed with the Fisher's least significant difference (LSD) multiple comparison method, each replicate considering 15 plants for treatment (n = 15). Data represent the average of four replicates ± standard error.
RNA extraction and cDNA synthesis
Total RNA was extracted from 100 mg of both shoot and root tissue sampling at T2 in hydroponics (Table 1) and at 66 days after sowing in open field for both genotypes. The Spectrum™ Plant Total RNA Kit (Sigma, St Luis; MO, USA) was used following the manufacturer’s protocol. Total RNA was then quantified with a Nanodrop1000 (Thermo Scientific, Nanodrop Products, Wilmington, DE, USA) and evaluated qualitatively by agarose gel electrophoresis. Then, cDNA was synthesized from 500 ng of total RNA mixed with 1 μL of 10 μM oligo-dT, as described by Manoli et al. (2012).
Gene selection for gene expression analysis
To better understand the molecular events occurring in response to N, a number of genes were selected according to their putative functions and/or to their transcriptional profiles in previous experiments (Li et al. 2023; Ravazzolo et al. 2021) (Table 2). In relation to their putative function they were grouped as those involved in SL production and signaling, those involved in the NO3− uptake and assimilation, and those contributing to amino acid, iron and sulfur transport and compartmentation.
Quantitative reverse transcription PCR (qRT-PCR)
Previous papers (Manoli et al. 2014, 2016; Trevisan et al. 2015, 2019; Ravazzolo et al. 2021, 2019) evidenced that a clear molecular regulation of the response to NO3− availability occurs already after few minutes/hour of NO3− provision and that it contributes to define the phenotype observed subsequently. For this reason, gene expression analyses was carried out at T2 under hydroponic conditions. For open field, leaf tissues were sampled at 66 days after sowing (Table 1). qRT-PCR was performed using the StepOne Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA USA) as described by Nonis et al. (2007). SYBR Green reagent (Applied Biosystems, Monza, Italy) was used, according to the manufacturer’s instructions. cDNA (2.5 ng) were used as template and three technical repetitions were performed on three biological repetitions. The absence of multiple products and primer dimers was confirmed by the melting curve analysis. Relative expression of the target gene was determined according to the Livak and Schmittgen (2001) method, using MEP (membrane protein PB1A10.07c, Zm00001d018359) as a housekeeping gene, according to Manoli et al. (2012). Primers were designed using the Primer3 web tool (version 4.0.0; http://bioinfo.ut.ee/primer3/; Rozen and Skaletsky, 2000). For statistical analysis, data was considered significant when p ≤ 0.05. by Student's t test for pairwise comparisons. The genes analyzed and the sequences of the relative primers used in qRT-PCR are reported in Table 2 and Online Resource 1.
Quantification of proteins through total Kjeldahl nitrogen (TKN), nitrate (NO3 −), sulfur (S), iron (Fe), potassium (K), magnesium (Mg) and manganese (Mn)
Since many of the genes considered for molecular analyses were involved in nutrient uptake and allocation, the total contents of proteins, nitrate (NO3−), iron (Fe), sulfur (S), potassium (K), magnesium (Mg) and manganese (Mn) were also determined. Samples were weighted as both fresh weight and dry weight after seven days at 60 °C and made to fine powder. Analyses were performed at the ‘LaChi’ laboratory of the University of Padua (Legnaro, Italy).
Crude protein content was determined by the analysis of the nitrogen content according to the total Kjeldahl nitrogen (TKN) (Hjalmarsson & Akesson 1983; AOAC 20th edition 2016.2001.11 rev 01) and calculated by multiplying the N content by a factor of 6.25 (Adler-Nissen 1986).
For the estimation of the anion NO3−, dry matter (200 mg) was extracted in water (50 mL) and filtered before analysis by ion chromatography (IC), as reported by Nicoletto et al. (2013).
For the estimation of iron (Fe), sulfur (S), potassium (K), magnesium (Mg) and manganese (Mn), the samples were first mineralized following the method reported by the AOAC (17th edition 2000.933.14), then the elements determination was conducted with inductively coupled plasma optical emission spectrometry (ICP-OES, Arcos II MV, Spectro A. I. GmbH, Kleve, Germany) (Mimmo et al. 2017).
Three biological replicates for each treatment and an ANOVA statistic test (p < 0.05) were performed with the Fisher's least significant difference (LSD) multiple comparison method each replicate considering 5 plants for treatment (n = 5). Data represent the average of three replicates ± standard error.
Results
WT and zmccd8 showed differences in biomass production
The phenotypic differences between WT and zmccd8 were first assessed in seedlings grown seven days in hydroponic conditions (T7) (Fig. 1). Both genotypes evidenced a dose-dependent response in terms of shoot length, with values increasing with the increase of the NO3− concentration (Fig. 1a). However, only slight differences were observed between genotypes at each concentration, with zmccd8 showing always longer shoot than the WT. On the contrary, the shoot dry weight was clearly lower compared to WT for all the nutritional regimes utilized (Fig. 1b). Both genotypes showed an increase in root elongation (Fig. 1a) and in dry weight (Fig. 1b) in response to increasing NO3− concentrations. Globally, these data evidence a clear distress of zmccd8 after 7 days of permanence in the nutrient solution in terms of biomass of the aerial part, apparently not depending on shoot elongation.
Length (cm) of primary roots and shoots (a), together with the average dry weight (DW, mg) per unit of roots and shoots (b) of wild-type maize seedlings (WT, black) and those of zmccd8 mutants (white) after seven (T7) days of growth in three different nutrient treatments: no N (0), KNO3 at 0.1 mM (0.1), and KNO3 at 1 mM (1). Error bars show the means of four biological replicates ± SE (standard error). An ANOVA statistic test with Fisher's least significant difference (LSD) multiple comparison method was performed, and values with the same lowercase letter are not significantly different from one another (P < 0.05)
Furthermore, in field conditions, regardless of N availability, the circumference (Fig. 2a) and height (Fig. 2b) of the stem, and the length of the internode (Fig. 2c) and leaves (Fig. 2e), and the number of leaves (Fig. 2d) showed significantly higher values for WT compared to zmccd8, suggesting a better growth capacity of the WT, as also confirmed by the phenotypic aspect (Fig. 2f).
Phenotypic analysis of wild-type (WT, black) and zmccd8 mutant plants (white) at 46, 56 and 66 days after sowing (das) under field conditions. The analysis included stem circumference (a), height (b), internode length (c), leaf number (d), and leaf length (e). Error bars represent the mean ± SE (n = 24). An ANOVA statistic test with Fisher's least significant difference (LSD) multiple comparison method was performed, and values with the same lowercase letter are not significantly different from one another (P < 0.05)
The zmccd8 mutant shows lower chlorophyll contents compared to WT
In N-deprived hydroponic solution, the CHL content was almost always significantly lower in both the first and the second leaves of zmccd8 compared to WT, with the only exception of the T10 time point when WT manifested a clear decrease in pigment accumulation, especially in the first leaf (Fig. 3a). At higher concentrations of NO3− (0.1 and 1 mM), the differences between the two genotypes were less marked in the first leaf, while in the second leaf zmccd8 always showed values significantly lower compared to the WT. Taken together, these data highlighted a lower global CHL content in the mutant compared to that of the WT.
Profiles in chlorophyll content (a) and anthocyanins levels (b) in the first and second leaves of wild-type maize plants (WT, solid lines with squares) and zmccd8 mutant seedlings (dotted lines with a circles) after three (T3), four (T4), six (T6), seven (T7), and ten (T10) days of growth in three different nutrient treatments: N-free (0), KNO3 at 0.1 mM, and KNO3 at 1.0 mM. Epidermal absorbance was quantified with the optical sensor DUALEX SCIENTIFIC + ™ (Force-A) at the hour indicated by each time point. Error bars represent the mean of six biological replicates ± SE. An ANOVA statistic test with Fisher's least significant difference (LSD) multiple comparison method was performed and different letters indicate different significance groups (P < 0.05)
In terms of the ANT content (Fig. 3b), after 10 days of N deprivation, WT showed a drastic accumulation of ANT in the first leaf. On the contrary, the mutant did not show a similar trend, maintaining an almost constant level of ANT at all time points. However, in the second leaf, the ANT content was slightly higher in the mutant compared to WT, at least at T3 and T6. For higher concentrations of NO3−, the increase in ANT accumulation observed for WT after 10 days was abolished and the trend observed for both genotypes was quite similar.
For field analyses, the third leaf was analyzed (Fig. 4a-b). CHL content was significantly higher in the WT, with values double compared to those measured in zmccd8 at each time-points. Similarly, the NBI resulted higher for WT at all the time-points, further supporting the hypothesis that the mutant zmccd8 has an altered response to N. On the contrary, ANT appeared always higher in zmccd8.
Leaf chlorophyll content (a), anthocyanin content (b), and Nitrogen Balance Index (c). Analyses were done on the same leaves for wild-type (WT, black) and mutant plants (zmccd8, white) at 46, 56 and 66 days after sowing (das) under field conditions. Epidermis absorbance was quantified with the optical sensor DUALEX SCIENTIFIC + ™ (Force-A) at the hour indicated by each time point. Values represent means ± SE (n = 24). Similar letters at the corresponding point within treatments are not significantly different (P < 0.05) by an ANOVA test with Fisher's least significant difference (LSD) multiple comparison method
The zmccd8 mutant showed an altered expression of genes involved in SL biosynthesis, signaling, and transport in hydroponics
To try to better deepen the observed differences between the two genotypes, the transcription of various genes involved in SL biosynthesis, signaling and transport (Table 2) was analyzed.
As expected, the expression of CCD8 in both leaves and roots of the mutant was essentially zero, while in WT it was up-regulated in response to N-starvation (Fig. 5). CCD7 expression was higher in root than in shoot for both genotypes, and slightly more abundant in zmccd8 plants compared to WT. MAX2, which encodes an F-box protein involved in SL signaling, and D53, which encodes a repressor of SL signaling, were transcribed in both shoots and roots of both genotypes and evidenced a slight decrease of expression in the mutant compared to the WT, possibly due to the absence of SLs. Finally, the putative SL transporter WBC33 was predominantly expressed in the shoot, where it was slightly induced in response to NO3− 1 mM in WT plants, but not in zmccd8. In roots of both, WBC33 transcript accumulation was clearly up-regulated in response to N starvation and it was more abundant in zmccd8. As far as PDR1 was concerned, its expression did not show significant differences between the two genotypes, except in the case of zmccd8 roots which showed a lower level of its expression at 0 mM NO3−. No evident trend of transcription was observed in response to nitrate.
Real-time qRT-PCR expression profiles of strigolactone (SL)-related genes in maize shoots and roots of wild-type (WT, black) and zmccd8 mutant seedlings (white) after two days (T2) of treatment with one of three nutrient regimes: N-free (0), KNO3 at 0.1 mM, or KNO3 at 1.0 mM. After 48 h of each treatment, the complete root and shoot systems were collected from each seedling (n = 4) and the relative mRNA levels for each gene were evaluated by qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Data are means ± SE for three biological replicates, * indicates differences between WT and zmccd8 at P < 0.05 by t test
The zmccd8 mutant is impaired in the ability to up-regulate genes encoding high-affinity transporters for nitrate acquisition in hydroponics
The transcription of two low-affinity NO3− transporters (NRT1.1 and NRT1.2) and one high-affinity NO3− transporters (NRT2.1) was analyzed (Fig. 6). NRT1.1 transcripts were detected in the shoot and root showing a slighter higher level of expression for WT. NRT1.2 was transcribed in both tissues, but predominantly in shoot were for WT and in the presence of NO3− 1 mM reached values more than double compared to zmccd8. Similar results were observed in root where at this nutritional condition the transcripts were four times higher than in zmccd8. In contrast, NRT2.1 was expressed mainly in root tissues and strongly induced at 0.1 mM NO3−, but it was then down regulated at 1 mM NO3−, as expected for a high-affinity NO3− transporter. In the mutant, this trend was less marked, with transcription values similar among the three nutritional conditions. This led to suggest that the mutant could be impaired in the ability to up-regulate the high-affinity system for NO3− acquisition.
Real-time qRT-PCR expression profiles of nitrogen (N)-related genes in maize shoots and roots of wild-type (WT, black) and zmccd8 mutant seedlings (white) after two days (T2) in one of three nutrient treatments: N-free treatment (0), KNO3 at 0.1 mM, or KNO3 at 1.0 mM. After 48 h of each treatment, complete root and shoot systems were collected from each seedling (n = 4) and the relative mRNA level for each gene was evaluated by qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Data are means ± SE for three biological replicates, * indicates differences between WT and zmccd8 at P < 0.05 by t test
The transcription of NR, encoding NO3− reductase (Fig. 6), was up-regulated in shoot in response to NO3− concentration and was always slightly higher in zmccd8 compared to WT. This behavior may lead to hypothesize a compensation for a lower NO3− uptake in the mutant. NR transcription was induced by NO3− also in roots, with no differences between genotypes.
As far as GS1 and GS2 transcription were concerned (Fig. 6), only minimal differences of transcription were noticed between genotypes and in response to NO3− provision in both roots and shoots. Regarding ASN3 (Fig. 6), in shoot at 0 and at 1 mM NO3− WT evidenced a significantly higher transcription compared to the mutant. Moreover, both genotypes showed a down-regulation of its transcription with increasing NO3− concentration. A similar regulation in response to NO3− concentration was also observed in roots, where zmccd8 showed a higher transcription of ASN3 only at 0 NO3−. As far as the ASN4 transcript accumulation was concerned, no significant differences between the two genotypes, nor in response to NO3− were observed (Fig. 6).
The transcription of genes encoding amino acid transporters was downregulated in zmccd8 in hydroponics
Five genes encoding amino acid (AA) transporters were selected as described in Table 2. AAAP10 was prevalently expressed in root, and it was slightly down-regulated in zmccd8 compared to the WT. The remaining four genes were expressed in both shoots and roots, but in most cases their expression was higher in WT, with differences generally more marked under no NO3− conditions (Fig. 7). These results led to hypothesize a global inhibition of AA transport/compartmentation in the zmccd8 mutant.
Real-time qRT-PCR expression profiles of amino acid (AA)-related genes in maize shoots and roots of wild-type (WT, black) and zmccd8 mutant seedlings (white) after two days (T2) in one of three nutrient treatments: N-free (0), KNO3 at 0.1 mM, or KNO3 at 1 mM. After 48 h of each treatment, complete root and shoot systems were collected from each seedling (n = 4) and relative mRNA levels for each gene were quantified by qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Data are means ± SE for three biological replicates, * indicates differences between WT and zmccd8 at P < 0.05 by t test
The zmccd8 mutant showed an altered transcription of genes encoding sulfate transporters in hydroponics
Three genes encoding sulfate transporters were identified as described above (Table 2). SULTR4 was predominantly expressed in roots where it showed increased accumulation with the increase of NO3− availability in the WT, while in zmccd8 this trend was absent, leading to strong differences in terms of gene transcription in correspondence of 1 mM NO3−. On the contrary, in shoot, SULTR4 was expressed more abundantly in zmccd8 than in WT at 1 mM NO3− (Fig. 8). SULTR5 was equally transcribed in shoot and root, showing slightly higher amounts of transcripts in WT shoot compared to those of zmccd8 and no significant differences in roots. As far as SULTR6 was concerned, it was highly transcribed in the shoot where it showed opposite profiles between the two genotypes depending on the NO3− concentrations. These data seem to suggest that S transport also could be affected in zmccd8.
Real-time qRT-PCR expression profiles of sulfur (S)-related genes in maize shoots and roots of wild-type (WT, black) and zmccd8 mutant seedlings (white) after two days (T2) in one of three nutrient treatments: N-free (0), KNO3 at 0.1 mM, or KNO3 at 1.0 mM. After 48 h of each treatment, complete root and shoot systems were collected from each seedling (n = 4) and relative mRNA for each gene was quantified by qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Data are means ± SE for three biological replicates, * indicates differences between WT and zmccd8 at P < 0.05 by t test
The zmccd8 mutant showed alterations in the transcription of genes involved in Fe distribution in hydroponics
NAS2, NAS4, NAS6, involved in nicotinamide biosynthesis, and VIT1 and VIT2, encoding vacuolar iron transporters (Table 2), showed a clear down expression in the zmccd8 (Fig. 9). NAS2 and NAS6 were predominantly transcribed in roots, while NAS4 was expressed both in leaves and roots. WT showed an increase of NAS4 transcript accumulation in shoot in response to NO3−, that was not observed for zmccd8. The same behavior was noticed for VIT 2 in root. Globally, these results seem to suggest that SLs could affect Fe chelation and transport within plant too.
Real-time qRT-PCR expression profiles of iron (Fe)-related genes in maize shoots and roots of wild-type (WT, black) and zmccd8 mutant seedlings (white) after two days (T2) in one of three nutrient treatments: N-free (0), KNO3 at 0.1 mM, or KNO3 at 1 mM. After 48 h of each treatment, complete root and shoot systems were collected from each seedling (n = 4) and relative mRNA for each gene was quantified by qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Data are means ± SE for three biological replicates, * indicates differences between WT and zmccd8 at P < 0.05 by t test
In the field, zmccd8 showed an altered expression of genes involved in SL and N metabolisms, transport and compartmentation of amino acids, Fe and S
The expression of previously selected genes (Table 2) was also evaluated in leaf samples from field (Fig. 10). As expected, no expression of CCD8 was detected in leaf of zmccd8, while transcription of CCD7, D53, MAX2, and WBC33 was significantly lower in the mutant compared to the WT (Fig. 10a). No significant variations were detected between WT and zmccd8 for PDR1.
Real-time qRT-PCR expression profiles for key genes in third leaves from wild-type (WT, black) and zmccd8 mutant plants (white) at 66 days after sowing (das) in the open field. Genes analyzed were related to strigolactone (SL) biosynthesis, signaling and transport (a), nitrogen (N) transport and metabolism (b), amino acid transport (c), sulfur (S) transport (d), and iron (Fe) transport and metabolism (e). After 66 days in the field, a sample from the third leaf was collected from all plants (n = 22 to 24) and relative mRNA levels for each gene were quantified using qRT-PCR. Transcript abundance is presented using mRNA levels normalized to MEP (Zm00001d018359, Manoli et al. 2012). Values represent means ± SE (n = 24), * indicates differences between WT and zmccd8 at P < 0.05 by t test
As far as genes involved in NO3− transport and assimilation were concerned (Fig. 10b), the transcription of NRT1.1 and NRT2.1 was almost absent in both WT and zmccd8 leaves, while that of NRT1.2 showed detectable level but not significant different among the two genotypes. NR showed the highest expression among this group, being slightly more transcribed in WT than in zmccd8. Similarly, even if with a global lower level, GS1 transcripts were a little more abundant in WT compared to zmccd8, while GS2 was equally little expressed in both genotypes. Finally, both ASN3 and ASN4 showed low level of expression in both genotypes, with ASN4 appearing more abundant in the zmccd8 mutant.
As fa as the genes encoding amino acid transporters (Fig. 10c) a significantly lower transcription of AAAP10, AAAP22, PTR2, and PTR5.6 in leaf of zmccd8 compared to that observed for the WT was noticed, while no significant variations were evidenced in AAAP29. A similar trend was also relieved for SULTR4 and SULTR6 encoding two sulfate transporters (Fig. 10d) and for NAS2, NAS4, NAS6, and VIT1 encoding three Nicotianamine Synthase and a vacuolar iron transporter, respectively (Fig. 10e). Altogether, these results confirm that SLs are important for the achievement of amino acids transport and Fe and S compartmentalization and led to hypothesize that these processes could take part to the complex interaction between SLs and N.
WT and zmccd8 show different content of proteins, NO3 −, S and Fe
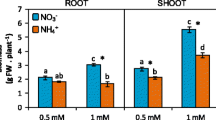
The whole plant total protein content was significantly higher in WT then in zmccd8 both in the presence and in the absence of NO3− when plants were grown in hydroponic. However, while WT in N-deprivation did not reduce the content of protein, zmccd8 showed a drastic decrease of it (Fig. 11a). Considering shoot, both genotypes showed a lower content of proteins under N-deficient conditions, even though the difference was slightly more marked for zmccd8 (Fig. 11b). As far as root were concerned, in N-starvation WT showed, on the contrary, a 50% increase of protein content, while zmccd8 still reduced it by more than half. A slightly higher content of protein for the WT was also observed in field (Fig. 11b). On the contrary, NO3− contents were always slightly higher in the tissues of zmccd8, both in hydroponic (Fig. 11a) and in field (Fig. 11b). Moreover, the supply of NO3− to the nutrient solution clearly stimulated a higher accumulation of the anion in all tissues of both genotypes.
Protein content obtained from total Kjeldahl nitrogen (TKN), together with nitrate (NO3−), sulfur (S), and iron (Fe) content as mg/g with respect to the dry weight (DW) in roots and shoots of wild-type (WT, black) and mutant maize seedlings (zmccd8, white) grown for 7 days in either complete nutrient solution (plus) or N-deficient solution (minus) (a), or content of protein, NO3−, S, and Fe with respect to the dry weight (DW) from the third leaf of WT or zmccd8 plants after 66 days in the open field (b). Values represent means ± SE (n = 5). Similar letters at corresponding points within the treatments are not significantly different (P < 0.05) by an ANOVA Fisher’s test
zmccd8 also showed lower values of Fe content compared to WT (Fig. 11a). Moreover, both genotypes showed an increase of it in N-deprivation. An identical trend was observed for shoots and root separately (Fig. 11a). In field, a slightly higher tissue content of Fe was observed for zmccd8 (Fig. 11b).
The whole plant S content almost duplicated in the WT grown at N-deficient conditions compared to plants grown with NO3− 1 mM, whereas in the case of zmccd8 it did not evidence significant changes (Fig. 11a). In shoot no differences were observed neither in response to nitrogen nor between genotypes (Fig. 11a). On the contrary, in roots WT almost triplicated the S content when grown in N-deprivation, whilst zmccd8 showed no changes in response to NO3− availability, with values identical to those measured for both genotypes when grown at 1 mM NO3−. In the field, third leaf of the mutant showed a significant lower S content compared to WT (Fig. 11b).
To assess the specificity of the effects of SLs on S and Fe compartmentation, the contents of K, Mg an Mn were also measured (Online Resource 2). No significant differences were detected between genotypes, except for Mg content in field leaf which resulted higher in the zmccd8 with respect to the WT.
Discussion
N deficiency is one of the most important abiotic factors reducing plant growth and crop yield (Zhang et al. 2020). In maize, N deficiency adversely affects growth, decreases shoot-to-root ratio (Steer & Harrigan 1986), and induces premature senescence (McConnell et al. 1995). Furthermore, remobilization of N from older to younger leaves and reproductive organs causes a leaf chlorosis characteristic of N deficiency (Sakuraba 2022). Levels of anthocyanin pigment also change (Ding et al. 2005). Since N is a central constituent of chlorophyll and of proteins associated with the photosynthetic apparatus, the availability of N readily affects photosynthesis, photosynthate partitioning, and leaf senescence (Xu et al. 2012). Actually, 70% of leaf N is accumulated in chloroplasts (Lu et al. 2023).
Multiple studies have demonstrated that N deficiency triggers strigolactone (SL) biosynthesis and exudation (Yoneyama et al. 2007; Sun et al. 2014; Ito et al. 2016; Yoneyama 2019; Ravazzolo et al. 2019, 2021), likely due to increasing transcript levels of SL biosynthetic genes, such as MAX3-MAX4 in Arabidopsis (Ito et al. 2016) and ZmCCD8 in maize (Ravazzolo et al. 2019, 2021). Orthologs of MAX3 encode the Carotenoid Cleavage Dioxygenase 7 (CCD7; Booker et al. 2004), whereas MAX4 orthologs encode the Carotenoid Cleavage Dioxygenase 8 (CCD8; Sorefan et al. 2003). The maize ZmCCD8 is a single-copy gene and a full loss of CCD8 function is achieved in the maize zmccd8::Ds mutant. Effects on maize architecture are pleiotropic but are relatively mild for apical dominance and bud outgrowth (Guan et al. 2012).
The present research was conducted both in hydroponic and field conditions and a comparative phenotypic and molecular analysis was directed to the zmccd8 mutant and wildtype (both in a B73 background) to better clarify the role of SLs in maize responses to N deficiency. NO3− and urea were provided as N source in hydroponic and in field, respectively. This choice is motivated by the fact that in soil urea is quickly hydrolyzed to NH4+ by ureases and NH4+ is then rapidly converted into NO3− (Pinton et al. 2016; Castaldelli et al. 2018).
Mutants deficient in SL biosynthesis or signaling are typically shorter with a modest increase in branching (Goulet & Klee 2010), as also observed in the present study (Fig. 2f). Our results highlight the central importance of SLs in acclimation of maize to N starvation (Figs. 1 and 2). In hydroponics zmccd8 mutant showed more pronounced deficiency symptoms in N deprivation compared to the WT (Figs. 1 and 2), slower growth, and lower chlorophyll content (Fig. 3). A stunted development and lower contents of chlorophyll were also observed in field regardless of N supply (Fig. 4). This part of the outcome was not surprising considering that SLs have a multi-faceted role in photosynthesis. By inhibiting activity of chlorophyll-degrading enzymes, SLs can alter the composition of photosynthetic pigments and thus the capacity of leaves for capturing light energy (Yamada et al. 2014). SLs also regulate the binding of chlorophyll to membrane proteins, maintain the stability of the chloroplast thylakoid membranes, and continuously enhance the photosynthetic capacity (Li et al. 2022).
To better understand molecular events underlying roles of SLs in the maize response to N-deprivation, we focused on genes described in Table 2, which were chosen according to their putative role and in light of their transcriptional profiles (Ravazzolo et al. 2021; Li et al. 2023), as described above. These included five sub-groups involved in: i) SL biosynthesis/signaling, ii) N transport and assimilation, iii) amino acid transport, iv) sulfate transport, and v) the transport and chelation of iron. Transcription of CCD8 was seriously compromised in zmccd8 mutants under all experimental conditions, confirming the expected phenotype (Figs. 5 and 10a). Transcription of all other genes in the SL sub-group was generally downregulated in shoots of the zmccd8 mutant. Levels of mRNA for putative SL transporters showed distinctive profiles with WBC33 being down-regulated in shoots, but up-regulated in roots, and PDR1 down regulated in roots, but only in N-deficiency. Results indicate that a lack of SL can impair the entire system for SL signalling and transport. In field conditions, all genes related to SL biosynthesis and signaling showed a lower level of expression if compared to hydroponics. This is probably due to a greater N availability than in hydroponics, since SL pathway is generally stimulated by N starvation (Fig. 10a).
The amount of transcripts of three nitrate transporters (NRT1.1, NRT1.2 and NRT2.1) was reduced in the zmccd8 mutant (Fig. 6, 10b). NRT1 belong to low affinity transporters (LATS) while NRT2 are part of the high affinity transport family (HATS), both being involved in root NO3− uptake and long-distance transport between and within plant organs (von Wittgenstein et al. 2014; Aluko et al. 2023). These results indicate a potentially lower efficiency for NO3− uptake and/or subsequent transport within mutant plants compared to wildtype. Among genes involved in the N reduction and assimilation, ASN3 appears particularly responsive to SLs, since its mRNAs levels were markedly decreased in the zmccd8 mutant shoots, mostly at low NO3− conditions. As the amino acid asparagine (Asn) is crucial for amino acid metabolism and usually serves as an N storage or transport metabolite (Schlüter et al. 2012), our results further support the importance of SLs for the correct functioning of N allocation processes. In addition, our data showed an overall lower expression of genes encoding putative amino acid transporters in the mutant compared to control plants (Figs. 7 and 10c), with transcripts for AAAP10, AAAP22 and PTR5.6 being most strongly reduced. These finding are coherent with those above, since AAAP proteins belong to the amino acid transporters (AAT) superfamily (Young et al. 1999). AAAP are mainly involved in regulating the long-distance transport of AA in the body of plants, mediating their transport across membrane structures and participating in a variety of other life processes, such as response to stresses (Hu et al. 2022). Sulphate transporter (SULTR) transcription was also affected in the mutant, suggesting that the uptake and translocation of sulphate may be impaired in the absence of SLs (Figs. 8 and 10d), thus compromising growth and plant performance that are strictly dependent on sulphur allocation in stressful conditions (Takahashi et al. 2011).
It must be noted that among the genes above mentioned, some genes displayed different transcription profiles among hydroponics and field conditions. For instance, in hydroponics CCD7 displayed a higher expression in the zmccd8 mutant if compared to WT in every nutritional conditions, probably as a compensation strategy due to the lack of expression of CCD8, while in the field this process was not visible because the plant was probably in a better nutritional status and did not need to increase the production of SLs. In addition, PTR2 and SULTR4 transcription were differently regulated in field and hydroponics, being more expressed in the WT in field conditions, while in hydroponics they displayed a higher expression in the zmccd8 mutant at 1 mM NO3−. This could rely to the differences availabilities of nutrients in soil compared to those provided in hydroponics, even though further work is needed to better assess and deepen these aspects.
Our results also showed an unequivocal down regulation of three Nicotianamine Synthase (NAS) genes in the zmccd8 mutant under almost all conditions tested, especially when NO3− levels were lowest (Figs. 9 and 10e), leading to suppose that SLs could also interfere with iron transport. A similar profile was also observed for VIT1, a gene encoding a vacuolar iron transporter. Fe is an essential component of several Fe-S proteins and is involved in physiological processes from photosynthesis to respiration (Kermeur et al. 2023). Grasses and grains have evolved a distinctive chelation-based “Strategy II” that mediates the uptake and transport of Fe in these species (Xu et al. 2022). This process requires biosynthesis of mugineic acids (MAs) and begins with methionine, progresses to nicotianamine, then produces and exudes the phytosiderophores (PS) (Nozoye et al. 2011). These PS not only promote uptake of Fe by roots, but also regulate formation of the Fe-PS complexes that translocate Fe to shoots (Nogiya et al. 2016). Nicotianamine itself is also responsible for long-distance translocation of Fe in stems (Zhou et al. 2013). Collective data led us to hypothesize that the acquisition and remobilization of iron in maize could at least in part depend on the biosynthesis of SLs. The link between Fe redistribution and SLs could also be related to the role of TOPLESS (TPL) and TPL-related proteins (TPL/TPRs), which regulate NAS4 during Fe deficiency (Brumbarova et al. 2015; Bai et al. 2020). In addition, the TPL and TPL/TPR proteins contribute to SL signaling through their physical interaction with SUPRESSOR OF MORE AXILLARY GROWTH2 1-LIKE (SMXL) (D53 in rice and maize) in complexes that regulate SL-responsive genes (Plant et al. 2021; Guan et al. 2022). In addition, in rice the presence of Fe is required for the NO3−-induced nuclear localization of NPL4, which in turn leads to SL signaling repression (Song et al. 2023). These finding reinforce the hypothesis of a connection between the N-Fe balance and SLs.
Although in hydroponics total protein content was minimally affected by N starvation of wildtype seedlings that were able to remobilize N from seeds (Fig. 11), the same was not observed for zmccd8 mutants. Instead, protein levels dropped markedly with N limitation when SLs were lacking and were especially pronounced in roots (Fig. 11a). In fact, wildtype roots at 0 NO3− had higher protein content than when grown with abundant N, whereas zmccd8 root protein was strongly decreased by N starvation. Contrarywise, wildtype plants always displayed lower levels of nitrates compared to zmccd8. This result led us to hypothesize that SLs contribute to an effective capacity for metabolizing/allocating/retaining root N under conditions of N deprivation. In fact, zmccd8 seems unable to correctly sense N deficiency and, as a consequence, to assimilate NO3− in organic compounds and remobilize these resources from other organs, as also confirmed by the molecular data. The proposed scenario is also supported by values of NBI in field experiments (Fig. 4) and by the lower protein content in the third leaf of zmccd8 compared to WT (Fig. 11b). The S content was also significantly higher in WT plants grown with limited N (Fig. 11) and most apparent in roots, where levels of S were almost four times greater. Instead, the SL-deficient zmccd8 mutants had significantly less S than did WT and this S content did not change with N availability (Fig. 11b). This intriguing contrast is consistent with the possibility that responses to N deprivation could enhance assimilation of S (the mechanisms of NO3− and sulfate assimilation in plastids having many commonalities) and that SL biosynthesis could be central in such process. While N directly affects photosynthetic efficiency of a plant, S indirectly affects photosynthesis by improving the NUE (Ahmad & Abdin 2000; Salvagiotti et al. 2009). An increase in S assimilation could thus potentially aids N deficiency, and similarities in their reductive metabolism could theoretically confer synergistic effects for scavenging limiting resources.
The Fe content of zmccd8 mutants was consistently less than that of wildtype plants regardless of N regime or tissue (Fig. 11a). However, levels were especially low in roots of zmccd8 mutants where Fe abundance was 2- to threefold less than in wildtype. An additional difference was the N-responsiveness of Fe accumulation by roots, which rose under conditions of N-starvation in wildtype plants, but did not in SL-deficient mutants. This behavior is comparable to that above described for sulfur and likely dependent on SLs (Fig. 11b).
A strong interdependence between responses to S and Fe has been thoroughly documented by Forieri et al. (2017) and Astolfi et al. (2021). Furthermore, a relationship between effects of N deficiency and homeostasis of S and Fe has been reported in Arabidopsis by two different groups (Chen et al. 2016; Brumbarova & Ivanov 2019).
K, Mg and Mn contents were also measured to assess the specificity of the effects observed for Fe and S in response to SLs (Online Resource 2). Results showed that K and Mn accumulation was not affected in the zmccd8, while Mg level was slightly higher in the field leaf sample of zmccd8, but statistically unaffected by the genotype in hydroponics. Agarwala and Mehrotra (1984) observed an antagonistic relationship between Fe and Mg in radishes, where nutrient accumulation was dependent upon whether there was a higher concentration of Mg or Fe in the soil. In addition, it was recently shown that rice plants treated with Mg tended to have smaller shoot Fe concentrations in the field, suggesting enhanced exclusion at the whole-plant level (Rajonandraina et al. 2023). Therefore, a compensation mechanism between Mg and Fe in the zmcced8 mutant could be hypothesized.
To conclude, collective evidence in the present study defines SLs as crucial factors for the physiological acclimation to nitrogen deficiency in maize. A list of molecular targets central to this regulation is also provided. Involvement can be direct and/or indirect and includes key genes for transport and distribution of NO3− and amino acids among plant tissues and organs. In addition, work here reveals a previously unrecognized association of sulfur and iron homeostasis with the interactions between nitrogen and strigolactones in maize. Our main conclusions are summarized in Fig. 12. Further work is needed to more precisely decipher the network linking N, S and Fe in the SL-mediated acclimation to nitrogen starvation. Nonetheless, data presented here demonstrate the importance of these constituents not only for their roles in plant nutrition but also for their contributions to stress tolerance. New insights are provided for SL functioning and possible applications in agriculture.
Data availability
All data generated or analyzed during this study are included in this published article.
Abbreviations
- SLs:
-
Strigolactones
- WT:
-
Wild type
- NUE:
-
Nitrogen use efficiency
- AMF:
-
Arbuscular mycorrhizal fungi
- PS:
-
Phytosiderophores
References
Abualia R, Ötvös K, Novák O et al (2022) Molecular framework integrating nitrate sensing in root and auxin-guided shoot adaptive responses. Proc Natl Acad Sci U S A 119:e2122460119. https://doi.org/10.1073/pnas.2122460119
Adler-Nissen J (1986) Enzymatic hydrolisis of food protein. Elsevier, New York
Agarwala SC, Mehrotra SC (1984) Iron-magnesium antagonism in growth and metabolism of radish. Plant Soil 80:355–361. https://doi.org/10.1007/BF02140042
Ahmad A, Abdin MZ (2000) Photosynthesis and its related physiological variables in the leaves of Brassica genotypes as influenced by sulphur fertilization. Physiol Plant 110:144–149. https://doi.org/10.1034/j.1399-3054.2000.110119.x
Akiyama K, Matsuzaki KI, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827. https://doi.org/10.1038/nature03608
Aluko OO, Kant S, Adedire OM et al (2023) Unlocking the potentials of nitrate transporters at improving plant nitrogen use efficiency. Front Plant Sci 14:1074839. https://doi.org/10.3389/fpls.2023.1074839
Anas M, Liao F, Verma KK et al (2020) Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol Res 53:47. https://doi.org/10.1186/s40659-020-00312-4
Asim M, Ullah Z, Oluwaseun A, Wang Q, Liu H (2020) Signalling overlaps between nitrate and auxin in regulation of the root system architecture: insights from the Arabidopsis thaliana. Int J Mol Sci 21:2880. https://doi.org/10.3390/ijms21082880
Astolfi S, Celletti S, Vigani G, Mimmo T, Cesco S (2021) Interaction between sulfur and iron in plants. Front Plant Sci 12:670308. https://doi.org/10.3389/fpls.2021.670308
Bai H, Cao H, Liu P et al (2020) Study of interaction between transcription factor ZmMYB153 and TPL/TPRs proteins. J Hebei Agric Univ 43:61–67. https://doi.org/10.13320/j.cnki.jauh.2020.0053
Barbier F, Fichtner F, Beveridge C (2023) The strigolactone pathway plays a crucial role in integrating metabolic and nutritional signals in plants. Nat Plants 9:1191–1200. https://doi.org/10.1038/s41477-023-01453-6
Bathaei A, Štreimikienė D (2023) A systematic review of agricultural sustainability indicators. Agriculture 13:241. https://doi.org/10.3390/agriculture13020241
Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14:1232–1238. https://doi.org/10.1016/j.cub.2004.06.061
Bouguyon E, Gojon A, Nacry P (2012) Nitrate sensing and signaling in plants. Semin Cell Dev Biol 23:648–654. https://doi.org/10.1016/j.semcdb.2012.01.004
Briat JF, Dubos C, Gaymard F (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20:33–40. https://doi.org/10.1016/j.tplants.2014.07.005
Brumbarova T, Ivanov R (2019) The nutrient response transcriptional regulome of Arabidopsis. iScience 19:358–368. https://doi.org/10.1016/j.isci.2019.07.045
Brumbarova T, Bauer P, Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20:124–133. https://doi.org/10.1016/j.tplants.2014.11.004
Castaldelli G, Colombani N, Tamburini E, Vincenzi F, Mastrocicco M (2018) Soil type and microclimatic conditions as drivers of urea transformation kinetics in maize plots. CATENA 166:200–208. https://doi.org/10.1016/j.catena.2018.04.009
Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X (2016) Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 26:640–646. https://doi.org/10.1016/j.cub.2015.12.066
Chiang CS, Stacey G, Tsay YF (2004) Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J Biol Chem 79:30150–30157. https://doi.org/10.1074/jbc.M405192200
Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154:1189–1190. https://doi.org/10.1126/science.154.3753.1189
Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1:e0011. https://doi.org/10.1199/tab.0011
Dechorgnat J, Francis KL, Dhugga KS, Rafalski JA, Tyerman SD, Kaiser BN (2018) root ideotype influences nitrogen transport and assimilation in maize. Front Plant Sci 9:531. https://doi.org/10.3389/fpls.2018.00531
Dechorgnat J, Francis KL, Dhugga KS, Rafalski JA, Tyerman SD, Kaiser BN (2019) Tissue and nitrogen-linked expression profiles of ammonium and nitrate transporters in maize. BMC Plant Biol 19:206. https://doi.org/10.1186/s12870-019-1768-0
Ding L, Wang KJ, Jiang GM et al (2005) Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Ann Bot 96:925–930. https://doi.org/10.1093/aob/mci244
Forieri I, Sticht C, Reichelt M, Gretz N, Hawkesford MJ, Malagoli M, Wirtz M, Hell R (2017) System analysis of metabolism and the transcriptome in Arabidopsis thaliana roots reveals differential co-regulation upon iron, sulfur and potassium deficiency. Plant Cell Environ 40:95–107. https://doi.org/10.1111/pce.12842
Gojon A (2017) Nitrogen nutrition in plants: rapid progress and new challenges. J Exp Bot 68:2457–2462. https://doi.org/10.1093/jxb/erx171
Gomez-Roldan V, Fermas S, Brewer PB et al (2008) Strigolactone inhibition of shoot branching. Nature 455:189–1894. https://doi.org/10.1038/nature07271
Goulet C, Klee HJ (2010) Climbing the branches of the strigolactones pathway one discovery at a time. Plant Physiol 154:493–496. https://doi.org/10.1104/pp.110.161026
Guan JC, Koch KE, Suzuki M et al (2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160:1303–1317. https://doi.org/10.1104/pp.112.204503
Guan JC, Li C, Flint-Garcia S, Suzuki M et al (2022) Maize domestication phenotypes reveal strigolactone networks coordinating grain size evolution with kernel-bearing cupule architecture. Plant Cell 35:1013–1037. https://doi.org/10.1093/plcell/koac370
Hjalmarsson S, Akesson R (1983) Modern Kjeldahl Procedure Int Lab 3:70–76
Hu L, Fan R, Wang P et al (2022) Identification, phylogenetic and expression analyses of the AAAP gene family in Liriodendron chinense reveal their putative functions in response to organ and multiple abiotic stresses. Int J Mol Sci 23:4765. https://doi.org/10.3390/ijms23094765
Huang Q, Wang M, Xia Z (2018) The SULTR gene family in maize (Zea mays L.): Gene cloning and expression analyses under sulfate starvation and abiotic stress. J Plant Physiol 220:24–33. https://doi.org/10.1016/j.jplph.2017.10.010
Ito S, Ito K, Abeta N, Takahashi R, Sasaki Y, Yajima S (2016) Effects of strigolactone signaling on Arabidopsis growth under nitrogen deficient stress condition. Plant Signal Behav 11:e1126031. https://doi.org/10.1080/15592324.2015.1126031
Kermeur N, Pédrot M, Cabello-Hurtado F (2023) Iron availability and homeostasis in plants: a review of responses, adaptive mechanisms, and signaling. Methods Molec Biol (Clifton, N.J.) 2642:49–81. https://doi.org/10.1007/978-1-0716-3044-0_3
Li S, Tian Y, Wu K et al (2018) Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560:595–600. https://doi.org/10.1038/s41586-018-0415-5
Li Y, Li S, Feng Q et al (2022) Effects of exogenous Strigolactone on the physiological and ecological characteristics of Pennisetum purpureum Schum. seedlings under drought stress. BMC Plant Biol 22:578. https://doi.org/10.1186/s12870-022-03978-y
Li C, Dong L, Durairaj J et al (2023) Maize resistance to witchweed through changes in strigolactone biosynthesis. Science 379:94–99. https://doi.org/10.1126/science.abq4775
Lin H, Wang R, Qian Q et al (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21:1512–1525. https://doi.org/10.1105/tpc.109.065987
Liu Y, Wu G, Zhao Y et al (2021) DWARF53 interacts with transcription factors UB2/UB3/TSH4 to regulate maize tillering and tassel branching. Plant Physiol 187:947–962. https://doi.org/10.1093/plphys/kiab259
Lu Y, Zhang X, Cui Y et al (2023) Response of different varieties of maize to nitrogen stress and diagnosis of leaf nitrogen using hyperspectral data. Sci Rep 13:5890. https://doi.org/10.1038/s41598-023-31887-z
Maghiaoui A, Gojon A, Bach L (2020) NRT1.1-centered nitrate signaling in plants. J Exp Bot 71:6226–6237. https://doi.org/10.1093/jxb/eraa36
Mallikarjuna MG, Thirunavukkarasu N, Sharma R et al (2020) Comparative transcriptome analysis of iron and zinc deficiency in maize (Zea mays L.). Plants (Basel) 9:1812. https://doi.org/10.3390/plants9121812
Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815. https://doi.org/10.1016/j.jplph.2012.01.019
Manoli A, Begheldo M, Genre A, Lanfranco L, Trevisan S, Quaggiotti S (2014) NO homeostasis is a key regulator of early nitrate perception and root elongation in maize. J Exp Bot 65:185–200. https://doi.org/10.1093/jxb/ert358
Manoli A, Trevisan S, Voigt B, Yokawa K, Baluška F, Quaggiotti S (2016) Nitric oxide-mediated maize root apex responses to nitrate are regulated by auxin and strigolactones. Front Plant Sci 6:1269. https://doi.org/10.3389/fpls.2015.01269
Marro N, Lidoy J, Chico MÁ et al (2022) Strigolactones: New players in the nitrogen-phosphorus signalling interplay. Plant Cell Environ 45:512–527. https://doi.org/10.1111/pce.14212
Marzec M, Melzer M (2018) Regulation of root development and architecture by strigolactones under optimal and nutrient deficiency conditions. Int J Mol Sci 19:1887. https://doi.org/10.3390/ijms19071887
McConnell JS, Glover RE, Vories ED et al (1995) Nitrogen fertilization and plant development of cotton as determined by nodes above white flower. J Plant Nutr 18:1027–1036. https://doi.org/10.1080/01904169509364958
Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58:2297–2306
Mimmo T, Tiziani R, Valentinuzzi F et al (2017) Selenium biofortification in Fragaria × ananassa: implications on strawberry fruits quality, content of bioactive health beneficial compounds and metabolomic profile. Front Plant Sci 8:1887. https://doi.org/10.3389/fpls.2017.01887
Mu X, Chen Q, Chen F, Yuan L, Mi G (2017) A RNA-seq analysis of the response of photosynthetic system to low nitrogen supply in maize leaf. Int J Mol Sci 18:2624. https://doi.org/10.3390/ijms18122624
Nicoletto C, Santagata S, Tosini F, Sambo P (2013) Qualitative and healthy traits of different Italian typical artichoke genotypes. CyTA-J Food 11:108–113. https://doi.org/10.1080/19476337.2012.700951
Nogiya M, Pandey RN, Singh B (2016) Physiological basis of iron chlorosis tolerance in rice (Oryza sativa) in relation to the root exudation capacity. J Plant Nutr 39:1536–1546. https://doi.org/10.1080/01904167.2016.1161786
Nozoye T, Nagasaka S, Kobayashi T et al (2011) Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J Biol Chem 286:5446–5454. https://doi.org/10.1074/jbc.M110.180026
O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate Transport, Sensing, and Responses in Plants. Mol Plant 9:837–856. https://doi.org/10.1016/j.molp.2016.05.004
Pan X, Zheng H, Zhao J, Xu Y, Li X (2016) ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 243:1407–1418. https://doi.org/10.1007/s00425-016-2479-5
Pinton R, Tomasi N, Zanin L (2016) Molecular and physiological interactions of urea and nitrate uptake in plants. Plant Signal Behav 11:e1076603. https://doi.org/10.1080/15592324.2015.1076603
Plant AR, Larrieu A, Causier B (2021) Repressor for hire! The vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol 231:963–973. https://doi.org/10.1111/nph.17428
Prinsi B, Espen L (2015) Mineral nitrogen sources differently affect root glutamine synthetase isoforms and amino acid balance among organs in maize. BMC Plant Biol 15:96. https://doi.org/10.1186/s12870-015-0482-9
Quaggiotti S, Ruperti B, Borsa P, Destro T, Malagoli M (2003) Expression of a putative high-affinity NO3- transporter and of an H+-ATPase in relation to whole plant nitrate transport physiology in two maize genotypes differently responsive to low nitrogen availability. J Exp Bot 54:1023–1031. https://doi.org/10.1093/jxb/erg106
Rajonandraina T, Ueda Y, Wissuwa M et al (2023) Magnesium supply alleviates iron toxicity-induced leaf bronzing in rice through exclusion and tissue-tolerance mechanisms. Front Plant Sci 14:1213456. https://doi.org/10.3389/fpls.2023.1213456
Ravazzolo L, Trevisan S, Manoli A, Boutet-Mercey SP, Perreau FO, Quaggiotti S (2019) The control of zealactone biosynthesis and exudation is involved in the response to nitrogen in maize root. Plant Cell Physiol 60:2100–2112. https://doi.org/10.1093/pcp/pcz108
Ravazzolo L, Boutet-Mercey S, Perreau F, Forestan C, Varotto S, Ruperti B, Quaggiotti S (2021) Strigolactones and auxin cooperate to regulate maize root development and response to nitrate. Plant Cell Physiol 62:610–623. https://doi.org/10.1093/pcp/pcab014
Sakakibara H, Shimizu H, Hase T, Yamazaki Y, Takao T, Shimonishi Y, Sugiyama T (1996) Molecular identification and characterization of cytosolic isoforms of glutamine synthetase in maize roots. J Biol Chem 271:29561–29568. https://doi.org/10.1074/jbc.271.47.29561
Sakuraba Y (2022) Molecular basis of nitrogen starvation-induced leaf senescence. Front Plant Sci 13:1013304. https://doi.org/10.3389/fpls.2022.1013304
Salvagiotti F, Castellarín JM, Miralles DJ, Pedrol HM (2009) Sulfur fertilization improves nitrogen use efficiency in wheat by increasing nitrogen uptake. Field Crop Res 113:170–177. https://doi.org/10.1016/j.fcr.2009.05.003
Schlüter U, Mascher M, Colmsee C, Scholz U, Bräutigam A, Fahnenstich H, Sonnewald U (2012) Maize source leaf adaptation to nitrogen deficiency affects not only nitrogen and carbon metabolism but also control of phosphate homeostasis. Plant Physiol 160:1384–1406. https://doi.org/10.1104/pp.112.204420
Sheng L, Deng L, Yan H et al (2014) A Genome-Wide Analysis of the AAAP Gene Family in Maize. J Proteomics Bioinform 7:23–33
Shindo M, Shimomura K, Yamaguchi S, Umehara M (2018) Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct 2:e00050. https://doi.org/10.1002/pld3.50
Sigalas PP, Buchner P, Thomas SG et al (2023) Nutritional and tissue-specific regulation of cytochrome P450 CYP711A MAX1 homologues and strigolactone biosynthesis in wheat. J Exp Bot 74:1890–1910. https://doi.org/10.1093/jxb/erad008
Sorefan K, Booker J, Haurogné K et al (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17:1469–1474. https://doi.org/10.1101/gad.256603
Steer BT, Harrigan EKS (1986) Rates of nitrogen supply during different developmental stages affect yield components of safflower (Carthamus tinctorius L.). Field Crops Research. https://doi.org/10.1016/0378-4290(86)90060-2
Sun H, Tao J, Liu S et al (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65:6735–6746. https://doi.org/10.1093/jxb/eru029
Sun J, Li W, Li C et al (2020) Effect of different rates of nitrogen fertilization on crop yield, soil properties and leaf physiological attributes in banana under subtropical regions of China. Front Plant Sci 11:613760. https://doi.org/10.3389/fpls.2020.613760
Sun H, Guo X, Zhu X et al (2023) Strigolactone and gibberellin signaling coordinately regulate metabolic adaptations to changes in nitrogen availability in rice. Mol Plant 16:588–598. https://doi.org/10.1016/j.molp.2023.01.009
Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62:157–184. https://doi.org/10.1146/annurev-arplant-042110-103921
Todd J, Screen S, Crowley J et al (2008) Identification and characterization of four distinct asparagine synthetase (AsnS) genes in maize (Zea mays L.). Plant Sci 175:799–808. https://doi.org/10.1016/j.plantsci.2008.08.004
Trevisan S, Manoli A, Begheldo M et al (2011) Transcriptome analysis reveals coordinated spatiotemporal regulation of hemoglobin and nitrate reductase in response to nitrate in maize roots. New Phytol 192:338–352. https://doi.org/10.1111/j.1469-8137.2011.03822.x
Trevisan S, Nonis A, Begheldo M et al (2012) Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ 35:1137–1155. https://doi.org/10.1111/j.1365-3040.2011.02478.x
Trevisan S, Trentin AR, Ghisi R, Masi A, Quaggiotti S (2019) Nitrate affects transcriptional regulation of UPBEAT1 and ROS localisation in roots of Zea mays L. Physiol Plant 166:794–811. https://doi.org/10.1111/ppl.12839
Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581:2290–2300. https://doi.org/10.1016/j.febslet.2007.04.047
Umehara M, Hanada A, Yoshida S et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200. https://doi.org/10.1038/nature07272
von Wittgenstein NJ, Le CH, Hawkins BJ, Ehlting J (2014) Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol Biol 14:11. https://doi.org/10.1186/1471-2148-14-11
Wani SH, Vijayan R, Choudhary M et al (2021) Nitrogen use efficiency (NUE): elucidated mechanisms, mapped genes and gene networks in maize (Zea mays L.). Physiol Molec Biol Plants 27:2875–2891. https://doi.org/10.1007/s12298-021-01113-z
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182. https://doi.org/10.1146/annurev-arplant-042811-105532
Xu J, Zhu X, Yan F, Zhu H, Zhou X, Yu F (2022) Identification of quantitative trait loci associated with iron deficiency tolerance in maize. Front Plant Sci 13:805247. https://doi.org/10.3389/fpls.2022.805247
Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240:399–408. https://doi.org/10.1007/s00425-014-2096-0
Yoneyama K (2019) How do strigolactones ameliorate nutrient deficiencies in plants? Cold Spring Harb Perspect Biol 11:a034686. https://doi.org/10.1101/cshperspect.a034686
Yoneyama K, Xie X, Kusumoto D et al (2007) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227:125–132. https://doi.org/10.1007/s00425-007-0600-5
Yoneyama K, Xie X, Kim HI et al (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235:1197–1207. https://doi.org/10.1007/s00425-011-1568-8
Young GB, Jack DL, Smith DW, Saier MH Jr (1999) The amino acid/auxin:proton symport permease family. Biochim Biophys Acta 1415:306–322. https://doi.org/10.1016/s0005-2736(98)00196-5
Yu P, Eggert K, von Wirén N, Li C, Hochholdinger F (2015) Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol 169:690–704. https://doi.org/10.1104/pp.15.00888
Yuan Z, Cao Q, Zhang K et al (2016) Optimal leaf positions for spad meter measurement in rice. Front Plant Sci 7:719. https://doi.org/10.3389/fpls.2016.00719
Zhang L, Liang Z, He X et al (2020) Improving grain yield and protein concentration of maize (Zea mays L.) simultaneously by appropriate hybrid selection and nitrogen management. Field Crops Res 249:107754. https://doi.org/10.1016/j.fcr.2020.107754
Zhou X, Li S, Zhao Q et al (2013) Genome-wide identification, classification and expression profiling of nicotianamine synthase (NAS) gene family in maize. BMC Genom 14:238. https://doi.org/10.1186/1471-2164-14-238
Acknowledgements
We are very grateful to Roberto Degan and Elisa Merlo for their essential help in the field analysis.
Funding
Open access funding provided by Università degli Studi di Padova within the CRUI-CARE Agreement. This work was supported by the University of Padova (DOR 2021 and BIRD 2020) and by European Project ‘AUTOSCREEN’ (LSHG-CT-2007–037897). Laura Ravazzolo was financed by a grant prot. BIRD211752-2021.
Author information
Authors and Affiliations
Contributions
Laura Ravazzolo, Leonardo Buzzicotti, Sara Trevisan and Silvia Quaggiotti performed the experiments. Laura Ravazzolo and Silvia Quaggiotti analyzed data and wrote the manuscript. Silvia Quaggiotti conceived and designed the project and obtained funds to support the project. Benedetto Ruperti and Serena Varotto contributed to funding the project. Karen E Koch and Jiahn Chou Guan provided seeds of the mutant zmccd8. Benedetto Ruperti, Serena Varotto, Karen E Koch and Jiahn-Chou Guan assisted with paper writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Weiming Shi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Quaggiotti, S., Buzzicotti, L., Koch, K.E. et al. Strigolactone roles in maize tolerance to low nitrogen involve shifts in acquisition and partitioning of protein, sulfur, and iron. Plant Soil (2024). https://doi.org/10.1007/s11104-024-06561-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11104-024-06561-6