Abstract
Aims
Revealing genetic diversity in a root nodulation symbiosis under field conditions is critical to understand the formation of ecological communities of organisms associated with hosts and the nitrogen cycle in natural ecosystems. However, our knowledge of the genetic diversity of bacterial mutualists on a local scale is still poor because of the assumption that the genetic diversity of mutualistic bacteria is constrained by their hosts.
Methods
We thoroughly investigated the genetic diversity of Frankia in a local forest stand. We collected root nodules from 213 Alnus hirsuta seedlings covering the spatial range of the continuous population, which means that Alnus individuals occurred in a relatively homogeneous distribution in a continuous forest. Then, a phylogenetic and diversity analysis was performed for the nifD-K IGS region, including global Frankia sequences from Alnus hosts.
Results
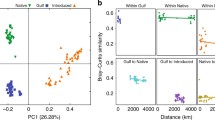
The genetic diversity of Frankia detected even on a local scale measured as high as that shown by previous studies conducted on local and regional scales. Moreover, a genetic structure analysis revealed a spatially mosaic-like distribution of genetic variation in Frankia despite the small spatial scale.
Conclusions
The genetic diversity and composition of bacterial mutualists are heterogeneous on a local scale. Our findings demonstrate that genetically different bacterial symbionts simultaneously interact with a single host population and interaction partnerships spatially vary. The standing variation could produce dynamic ecological and evolutionary outcomes in a heterogeneous forest ecosystem.





Similar content being viewed by others
Data availability
The sequence data are deposited at DDBJ with accession numbers of LC482655–LC482672 and LC528956–LC529138.
References
Ager AA, Heilman PE, Stettler RF (1993) Genetic variation in red alder (Alnus rubra) in relation to native climate and geography. Can J For Res 23:1930–1939. https://doi.org/10.1139/x93-243
Anderson MD, Ruess RW, Myrold DD, Taylor DL (2009) Host species and habitat affect nodulation by specific Frankia genotypes in two species of Alnus in interior Alaska. Oecologia 160:619–630. https://doi.org/10.1007/s00442-009-1330-0
Anderson MD, Taylor DL, Ruess RW (2013) Phylogeny and assemblage composition of Frankia in Alnus tenuifolia nodules across a primary successional sere in interior Alaska. Mol Ecol 22:3864–3877. https://doi.org/10.1111/mec.12339
Archetti M, Úbeda F, Fudenberg D, Green J, Pierce NE, Yu DW (2011) Let the right one in: a microeconomic approach to partner choice in mutualisms. Am Nat 177:75–85. https://doi.org/10.1086/657622
Arveby AS, Huss-Danell K (1988) Presence and dispersal of infective Frankia in peat and meadow soils in Sweden. Biol Fert Soils 6:39–44. https://doi.org/10.1007/BF00257918
Baker DD (1987) Relationships among pure cultured strains of Frankia based on host specificity. Physiol Plantarum 70:245–248. https://doi.org/10.1111/j.1399-3054.1987.tb06139.x
Ballhorn DJ, Elias JD, Balkan MA, Fordyce RF, Kennedy PG (2017) Colonization by nitrogen-fixing Frankia bacteria causes short-term increases in herbivore susceptibility in red alder (Alnus rubra) seedlings. Oecologia 184:497–506. https://doi.org/10.1007/s00442-017-3888-2
Barrett LG, Broadhurst LM, Thrall PH (2012) Geographic adaptation in plant-soil mutualisms: tests using Acacia spp. and rhizobial bacteria. Funct Ecol 26:457–468. https://doi.org/10.1111/j.1365-2435.2011.01940.x
Ben Tekaya S, Guerra T, Rodriguez D, Dawson JO, Hahn D (2018) Frankia diversity in host plant root nodules is independent of abundance or relative diversity of Frankia populations in corresponding rhizosphere soils. Appl Environ Microb 84:1–11. https://doi.org/10.1128/AEM.02248-17
Benson DR, Dawson JO (2007) Recent advances in the biogeography and genecology of symbiotic Frankia and its host plants. Physiol Plantarum 130:318–330. https://doi.org/10.1111/j.1399-3054.2007.00934.x
Benson DR, Hanna D (1983) Frankia diversity in an alder stand as estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis of whole-cell proteins. Can J Botany 61:2919–2923. https://doi.org/10.1139/b83-325
Berry AM, Mendoza-Herrera A, Guo Y et al (2011) New perspectives on nodule nitrogen assimilation in actinorhizal symbioses. Funct Plant Biol 38:645–652. https://doi.org/10.1071/FP11095
Burleigh SH, Dawson JO (1995) Spores of Frankia strain HFPCcl3 nodulate Casuarina equisetifolia after passage through the digestive tracts of captive parakeets (Melopsittacus undulatus). Can J Botany 73:1527–1530. https://doi.org/10.1139/b95-165
Caldwell BE (1966) Inheritance of a strain-specific ineffective nodulation in soybeans. Crop Sci 6:427–428. https://doi.org/10.2135/cropsci1966.0011183X000600050010x
Chaia EE, Sosa MC, Raffaele E (2012) Vertebrate faeces as sources of nodulating Frankia in Patagonia. Symbiosis 56:139–145. https://doi.org/10.1007/s13199-012-0169-z
Chen Z, Li J (2004) Phylogenetics and biogeography of Alnus (Betulaceae) inferred from sequences of nuclear ribosomal DNA ITS region. Int J Plant Sci 165:325–335. https://doi.org/10.1086/382795
Clawson ML, Caru M, Benson DR (1998) Diversity of Frankia strains in root nodules of plants from the families Elaeagnaceae and Rhamnaceae. Appl Environ Microb 64:3539–3543. https://doi.org/10.5301/HIP.2012.9281
Clawson ML, Gawronski J, Benson DR (1999) Dominance of Frankia strains in stands of Alnus incana subsp. rugosa and Myrica pensylvanica. Can J Botany 77:1203–1207. https://doi.org/10.1139/b99-070
Cunnings A, Johnson E, Martin Y (2016) Fluvial seed dispersal of riparian trees: transport and depositional processes. Earth Surf Proc Land 41:615–625. https://doi.org/10.1002/esp.3850
Dean J, Mescher M, De Moraes C (2014) Plant dependence on rhizobia for nitrogen influences induced plant defenses and herbivore performance. Int J Mol Sci 15:1466–1480. https://doi.org/10.3390/ijms15011466
Dillon JT, Baker D (1982) Variations in nitrogenase activity among pure-cultured Frankia strains tested on actinorhizal plants as an indication of symbiotic compatibility. New Phytol 92:215–219. https://doi.org/10.1111/j.1469-8137.1982.tb03378.x
Du D, Baker DD (1992) Actinorhizal host-specificity of Chinese Frankia strains. Plant Soil 144:113–116. https://doi.org/10.1007/BF00018851
Ellison AM, Bank MS, Clinton BD et al (2005) Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front Ecol Environ 3:479–486. https://doi.org/10.1890/1540-9295(2005)003[0479:LOFSCF]2.0.CO;2
Ellison AM, Barker-Plotkin AA, Foster DR, Orwig DA (2010) Experimentally testing the role of foundation species in forests: the Harvard Forest Hemlock Removal Experiment. Methods Ecol Evol 1:168–179. https://doi.org/10.1111/j.2041-210x.2010.00025.x
Ferriere R, Bronstein JL, Rinaldi S, Law R, Gauduchon M (2002) Cheating and the evolutionary stability of mutualisms. P Roy Soc Lond B Bio 269:773–780. https://doi.org/10.1098/rspb.2001.1900
Hahn D, Starrenburg MJC, Akkermans ADL (1988) Variable compatibility of cloned Alnus glutinosa ecotypes against ineffective Frankia strains. Plant Soil 107:233–243. https://doi.org/10.1007/BF02370552
Hayashi M, Saeki Y, Haga M, Harada K, Kouchi H, Umehara Y (2012) Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breeding Sci 61:544–553. https://doi.org/10.1270/jsbbs.61.544
He XH, Chen LG, Hu XQ, Asghar S (2004) Natural diversity of nodular microsymbionts of Myrica rubra. Plant Soil 262:229–239. https://doi.org/10.1023/B:PLSO.0000037045.42440.1d
Heath KD, Stinchcombe JR (2014) Explaining mutualism variation: a new evolutionary paradox? Evolution 68:309–317. https://doi.org/10.1111/evo.12292
Heath KD, Tiffin P (2007) Context dependence in the coevolution of plant and rhizobial mutualists. P Roy Soc B Biol Sci 274:1905–1912. https://doi.org/10.1098/rspb.2007.0495
Hoeksema JD, Kummel M (2003) Ecological persistence of the plant-mycorrhizal mutualism: a hypothesis from species coexistence theory. Am Nat 162:S40–S50. https://doi.org/10.1086/378644
Hooker JE, Wheeler CT (1987) The effectivity of Frankia for nodulation and nitrogen fixation in Alnus rubra and A. glutinosa. Physiol Plantarum 70:333–341. https://doi.org/10.1111/j.1399-3054.1987.tb06152.x
Huguet V, Batzli JM, Zimpfer JF, Normand P, Dawson JO, Fernandez MP (2001) Diversity and specificity of Frankia strains in nodules of sympatric Myrica gale, Alnus incana, and Shepherdia canadensis determined by rrs gene polymorphism. Appl Environ Microb 67:2116–2122. https://doi.org/10.1128/AEM.67.5.2116-2122.2001
Huss-Danell K, Uliassi D, Renberg I (1997) River and lake sediments as sources of infective Frankia (Alnus). Plant Soil 197:35–39. https://doi.org/10.1023/A:1004268931699
Jiabin H, Zheying Z, Guanxiong C, Huichang L (1985) Host range of Frankia endophytes. Plant Soil 87:61–65. https://doi.org/10.1007/BF02277648
Kagiya S, Yasugi M, Kudoh H, Nagano AJ, Utsumi S (2018) Does genomic variation in a foundation species predict arthropod community structure in a riparian forest? Mol Ecol 27:1284–1295. https://doi.org/10.1111/mec.14515
Kennedy PG, Weber MG, Bluhm AA (2010) Frankia bacteria in Alnus rubra forests: genetic diversity and determinants of assemblage structure. Plant Soil 335:479–492. https://doi.org/10.1007/s11104-010-0436-9
Khan A, Myrold DD, Misra AK (2007) Distribution of Frankia genotypes occupying Alnus nepalensis nodules with respect to altitude and soil characteristics in the Sikkim Himalayas. Physiol Plantarum 130:364–371. https://doi.org/10.1111/j.1399-3054.2006.00872.x
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
King RA, Ferris C (1998) Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Mol Ecol 7:1151–1161. https://doi.org/10.1046/j.1365-294x.1998.00432.x
Kucho K, Hay A, Normand P (2010) The determinants of the actinorhizal symbiosis. Microbes Environ 25:241–252. https://doi.org/10.1264/jsme2.ME10143
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lawrence DB, Schoenike RE, Quispel A, Bond G (1967) The role of Dryas drummondii in vegetation development following ice recession at Glacier Bay, Alaska, with special reference to its nitrogen fixation by root nodules. J Ecol 55:793–813. https://doi.org/10.2307/2258426
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Markham JH (2008) Variability of nitrogen-fixing Frankia on Alnus species. Botany 86:501–510. https://doi.org/10.1139/B08-023
Miller RW, Sirois JC (1982) Relative efficacy of different alfalfa cultivar-Rhizobium meliloti strain combinations for symbiotic nitrogen fixation. Appl Environ Microb 43:764–768. http://www.ncbi.nlm.nih.gov/pubmed/16345986
Mirza BS, Welsh A, Rasul G, Rieder JP, Paschke MW, Hahn D (2009) Variation in Frankia populations of the Elaeagnus host infection group in nodules of six host plant species after inoculation with soil. Microb Ecol 58:384–393. https://doi.org/10.1007/s00248-009-9513-0
Mishra AK, Singh PK, Singh P, Singh A, Singh SS, Srivastava A, Srivastava AK, Sarma HK (2015) Phylogeny and evolutionary genetics of Frankia strains based on 16S rRNA and nifD-K gene sequences. J Basic Microb 55:1013–1020. https://doi.org/10.1002/jobm.201400914
Normand P, Orso S, Cournoyer B et al (1996) Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol 46:1–9. https://doi.org/10.1099/00207713-46-1-1
Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, Couloux A, Cournoyer B, Cruveiller S, Daubin V, Demange N, Francino MP, Goltsman E, Huang Y, Kopp OR, Labarre L, Lapidus A, Lavire C, Marechal J, Martinez M, Mastronunzio JE, Mullin BC, Niemann J, Pujic P, Rawnsley T, Rouy Z, Schenowitz C, Sellstedt A, Tavares F, Tomkins JP, Vallenet D, Valverde C, Wall LG, Wang Y, Medigue C, Benson DR (2007) Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res 17:7–15. https://doi.org/10.1101/gr.5798407
Nouioui I, Ghodhbane-Gtari F, Fernandez MP, Boudabous A, Normand P, Gtari M (2014) Absence of cospeciation between the uncultured Frankia microsymbionts and the disjunct actinorhizal Coriaria species. Biomed Res Int 2014:1–9. https://doi.org/10.1155/2014/924235
Nyeko P, Edwards-Jones G, Day RK (2002) Population dynamics of herbivorous insects and potential arthropod natural enemies on Alnus species in Kabale district, Uganda. Agrofor Syst 56:213–224. https://doi.org/10.1023/A:1021376414975
Oksanen J, Blanchet FG, Friendly M et al (2019). Vegan: community ecology package. R Package Version 2.5–6. Retrieved from https://github.com/vegandevs/vegan
Pahua VJ, Stokes PJN, Hollowell AC, Regus JU, Gano-Cohen KA, Wendlandt CE, Quides KW, Lyu JY, Sachs JL (2018) Fitness variation among host species and the paradox of ineffective rhizobia. J Evolution Biol 31:599–610. https://doi.org/10.1111/jeb.13249
Paschke MW, Dawson JO (1993) Avian dispersal of Frankia. Can J Botany 71:1128–1131. https://doi.org/10.1139/b93-132
Peakall R, Smouse PE. (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pebesma E, Bivand R, Rowlingson B et al (2018). sp: Classes and methods for spatial data. R Package Version 1.3–1. Retrieved from https://github.com/edzer/sp
Pokharel A, Mirza BS, Dawson JO, Hahn D (2011) Frankia populations in soil and root nodules of sympatrically grown Alnus taxa. Microb Ecol 61:92–100. https://doi.org/10.1007/s00248-010-9726-2
Põlme S, Bahram M, Kõljalg U, Tedersoo L (2014) Global biogeography of Alnus-associated Frankia actinobacteria. New Phytol 204:979–988. https://doi.org/10.1111/nph.12962
Pozzi AC, Bautista-Guerrero HH, Nouioui I, Cotin-Galvan L, Pepin R, Fournier P, Menu F, Fernandez MP, Herrera-Belaroussi A (2015) In-planta sporulation phenotype: a major life history trait to understand the evolution of Alnus-infective Frankia strains. Environ Microbiol 17:3125–3138. https://doi.org/10.1111/1462-2920.12644
Pozzi AC, Roy M, Nagati M, Schwob G, Manzi S, Gardes M, Moreau PA, Fernandez MP (2018) Patterns of diversity, endemism and specialization in the root symbiont communities of alder species on the island of Corsica. New Phytol 219:336–349. https://doi.org/10.1111/nph.14996
Pozzi AC, Bautista-Guerrero HH, Abby SS, Herrera-Belaroussi A, Abrouk D, Normand P, Menu F, Fernandez MP (2018a) Robust Frankia phylogeny, species delineation and intraspecies diversity based on multi-locus sequence analysis (MLSA) and single-locus strain typing (SLST) adapted to a large sample size. Syst Appl Microbiol 41:311–323. https://doi.org/10.1016/j.syapm.2018.03.002
Pozzi AC, Roy M, Nagati M, Schwob G, Manzi S, Gardes M, Moreau PA, Fernandez MP (2018b) Patterns of diversity, endemism and specialization in the root symbiont communities of alder species on the island of Corsica. New Phytol 219:336–349. https://doi.org/10.1111/nph.14996
Prat D (1989) Effects of some pure and mixed Frankia strains on seedling growth in different Alnus species. Plant Soil 113:31–38. https://doi.org/10.1007/BF02181918
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Reddell P, Spain AV (1991) Transmission of infective Frankia (Actinomycetales) propagules in casts of the endogeic earthworm Pontoscolex corethrurus (Oligochaeta: Glossoscolecidae). Soil Biol Biochem 23:775–778. https://doi.org/10.1016/0038-0717(91)90148-D
Ren BQ, Xiang XG, Chen ZD (2010) Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Mol Ecol Resour 10:594–605. https://doi.org/10.1111/j.1755-0998.2009.02815.x
Ridgway KP, Marland LA, Harrison AF, Wright J, Young JPW, Fitter AH (2004) Molecular diversity of Frankia in root nodules of Alnus incana grown with inoculum from polluted urban soils. FEMS Microbiol Ecol 50:255–263. https://doi.org/10.1016/j.femsec.2004.07.002
Robinson KO, Beyene DA, van Berkum P, Knight-Mason R, Bhardwaj HL (2000) Variability in plant-microbe interaction between Lupinus lines and Bradyrhizobium strains. Plant Sci 159:257–264. https://doi.org/10.1016/S0168-9452(00)00345-9
Roy M, Pozzi AC, Gareil R et al (2017) Alder and the golden fleece: high diversity of Frankia and ectomycorrhizal fungi revealed from Alnus glutinosa subsp. barbata roots close to a tertiary and glacial refugium. PeerJ 5:e3479. https://doi.org/10.7717/peerj.3479
Sellstedt A, Huss-Danell K, Ahlqvist A (1986) Nitrogen fixation and biomass production in symbioses between Alnus incana and Frankia strains with different hydrogen metabolism. Physiol Plantarum 66:99–107. https://doi.org/10.1111/j.1399-3054.1986.tb01240.x
Simonet P, Le Thi N, Moiroud A, Bardin R (1989) Diversity of Frankia strains isolated from a single alder stand. Plant Soil 118:13–22. https://doi.org/10.1007/BF02232786
Simonet P, Bosco M, Chapelon C, Moiroud A, Normand P. (1994) Molecular characterization of Frankia microsymbionts from spore-positive and spore-negative nodules in a natural alder stand. Appl Environ Microbiol 60:1335–1341. https://aem.asm.org/content/60/4/1335/article-info
Thamer S, Schädler M, Bonte D, Ballhorn DJ (2011) Dual benefit from a belowground symbiosis: nitrogen fixing rhizobia promote growth and defense against a specialist herbivore in a cyanogenic plant. Plant Soil 341:209–219. https://doi.org/10.1007/s11104-010-0635-4
Torrey JG (1987) Endophyte sporulation in root nodules of actinorhizal plants. Physiol Plantarum 70:279–288. https://doi.org/10.1111/j.1399-3054.1987.tb06145.x
Vanden Heuvel BD, Benson DR, Bortiri E, Potter D (2004) Low genetic diversity among Frankia spp. strains nodulating sympatric populations of actinorhizal species of Rosaceae, Ceanothus (Rhamnaceae) and Datisca glomerata (Datiscaceae) west of the Sierra Nevada (California). Can J Microbiol 50:989–1000. https://doi.org/10.1139/w04-079
Wheeler CT, Akkermans ADL, Berry AM (2008) Frankia and actinorhizal plants: a historical perspective. In: Pawlowski K, Newton WE, eds. Nitrogen-fixing actinorhizal symbioses. Dordrecht: Springer US, 1–24
Wickneswari R, Norwati M (1993) Genetic diversity of natural-populations of Acacia auriculiformis. Aust J Bot 41:65–77. https://doi.org/10.1071/BT9930065
Wilcox DA, Cowan DA (2016) Diversity of Frankia in root nodules of six Morella sp. from the Cape flora of South Africa. Plant Soil 406:375–388. https://doi.org/10.1007/s11104-016-2881-6
Yamakawa T, Hussain AKMA, Ishizuka J (2003) Soybean preference for Bradyrhizobium japonicum for nodulation. Soil Sci Plant Nutr 49:835–841. https://doi.org/10.1080/00380768.2003.10410345
Acknowledgments
We thank Rei Sakai, Michio Ishihara, Takashi Hara, Yoshiaki Takahashi, Masayuki Watarai, Wataru Mamiya, Yuri Yabuhara, and members of Uryu Experimental Forest, Hokkaido University, for their field assistance.
Funding
This research was supported by Grant-in-Aid for JSPS Research Fellow to SK (16 J03194), Young Scientists A (16H0617908), and Scientific Research B (19H0297409) to SU.
Author information
Authors and Affiliations
Contributions
SK and SU designed and conducted the investigation, performed the molecular analysis, analyzed the data, and wrote the manuscript.
Corresponding author
Additional information
Responsible Editor: Katharina Pawlowski.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 333 kb)
Rights and permissions
About this article
Cite this article
Kagiya, S., Utsumi, S. Spatial heterogeneity in genetic diversity and composition of bacterial symbionts in a single host species population. Plant Soil 452, 513–527 (2020). https://doi.org/10.1007/s11104-020-04583-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-020-04583-4