Abstract
Aims
The objective of this research was to develop a three-dimensional (3D) rhizosphere modeling capability for plant-soil interactions by integrating plant biophysics, water and ion uptake and release from individual roots, variably saturated flow, and multicomponent reactive transport in soil.
Methods
We combined open source software for simulating plant and soil interactions with parallel computing technology to address highly-resolved root system architecture (RSA) and coupled hydrobiogeochemical processes in soil. The new simulation capability was demonstrated on a model grass, Brachypodium distachyon.
Results
In our simulation, the availability of water and nutrients for root uptake is controlled by the interplay between 1) transpiration-driven cycles of water uptake, root zone saturation and desaturation; 2) hydraulic redistribution; 3) multicomponent competitive ion exchange; 4) buildup of ions not taken up during kinetic nutrient uptake; and 5) advection, dispersion, and diffusion of ions in the soil. The uptake of water and ions by individual roots leads to dynamic, local gradients in ion concentrations.
Conclusion
By integrating the processes that control the fluxes of water and nutrients in the rhizosphere, the modeling capability we developed will enable exploration of alternative RSAs and function to advance the understanding of the coupled hydro-biogeochemical processes within the rhizosphere.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rhizosphere is a complex interaction zone for hydrological and biogeochemical processes characterized by large gradients in chemical and biological concentrations and physical properties along the root-soil interface (Helliwell et al. 2017). Root system and rhizosphere processes govern ecosystem plant water and nutrient use efficiency. Representations of these processes in numerical models are critical because they profoundly impact global carbon stocks and vegetation feedbacks (Finzi et al. 2015). However, the rhizosphere often has been studied independently by soil physicists, chemists, microbiologists, and plant physiologists (York et al. 2016). The coupled impacts of hydrologic, geochemical and biological processes on nutrient and carbon flow through roots and soil, have been identified as a persistent critical knowledge gap (Matamala and Stover 2013; Vereecken et al. 2016). Nutrient availability is critical for plant growth and productivity (Lopez-Bucio et al. 2003). Hinsinger et al. (2008) emphasized that the multicomponent, reactive transport models developed in geochemistry should be used to account for the numerous concurrent biogeochemical processes that interact with plant nutrient availability. Here, the term “multicomponent reactive transport” is taken to mean that multiple reactions can share the same ion. This can lead to interactions between ions and competition for common ions that systematically control concentrations. For example, changes in the pH or the aqueous concentration of an individual exchangeable cation systematically affects the concentration of all exchangeable cations (Chung and Zasoski 1994). This differs from some nutrient uptake models which may have multiple nutrients, but without interactions.
Root system architecture (RSA) is an important control on water and nutrient uptake from the soil. Numerous models of varying complexity have been developed to study the hydrologic processes in the rhizosphere including the two-way feedback between the root xylem and the soil. Some models use a circuit analogy to simulate the flow of water through the network of xylem conduits (Christoffersen et al. 2016; Garcia-Tejera et al. 2017; Manoli et al. 2017; Sperry et al. 1998). The soil compartments in these models are often simplified shells surrounding one-dimensional (1D) root layers. Others are fully coupled root and soil systems, either in one (Amenu and Kumar 2008) or three dimensions considering explicit root architecture (Doussan et al. 2006; Dunbabin et al. 2013; Javaux et al. 2008; Leitner et al. 2014; Manoli et al. 2014; Nietfeld and Prenzel 2015; Tournier et al. 2015). Three-dimensional (3D) models can better resolve the spatial distribution of water and nutrient resulting from interactions between individual roots and the local soil hydrobiogeochemistry (Garrigues et al. 2006; Manoli et al. 2014). On the other hand, simulations of nutrient dynamics are mainly limited to one and two dimensions (Espeleta et al. 2017; Mollier et al. 2008; Nietfeld and Prenzel 2015), or 3D non-reactive nutrient uptake, explicitly taking into account the geometry of the plant root system (Clausnitzer and Hopmans 1994; Dunbabin et al. 2004; Dunbabin et al. 2006; Leitner et al. 2010c; Simunek and Hopmans 2009; Tournier et al. 2015). The model by Gérard et al. (2017) was the first to couple a root system architecture model with a multicomponent reactive transport model using a macroscopic approach. They demonstrated their model through an application to P acquisition in an alkaline soil. However, they also ignored the presence of water flow in root and soil in their simulations. There is a need for 3D coupled plant root-soil system simulation capabilities that integrate 1) diel cycles of transpiration-driven root water uptake, 2) water uptake and release by individual roots in a RSA, 3) nutrient/ion uptake by individual roots, 4) variably saturated soil-root xylem water flow, 5) advection, dispersion, and diffusion of ions in the soil, and 6) multicomponent reactions.
Knowledge and experience from different disciplines are needed to collectively address critical knowledge gaps in our understanding of soil-plant interactions in the rhizosphere (Vereecken et al. 2016; Zhu et al. 2016). Here, we describe a new modeling capability that is intended to facilitate plant-soil hydrobiogeochemistry targeted by the plant and soil science modeling communities. The new capability exploits the availability of process-rich open source 3D simulation capabilities for soil water, reactive transport and RSA, as well as high-performance computing technologies. Our modeling capability can be used to characterize root functions and RSA, study and test interactions with soil hydrobiogeochemical processes, and systematically investigate the results of alternative RSAs, soil properties, water and nutrient dynamics in plant-soil systems within hours of wall clock time. Specifically, we describe the integration of process models to address the aforementioned need for 3D coupled plant root-soil system simulation capabilities using open source software. The computational kernel for the coupled hydrobiogeochemistry is designed for execution on desktop to massively parallel computing architectures. We report case studies of root water and nutrient uptake for Brachypodium distachyon (B. distachyon), a C3 model grass for food and bioenergy crops (Brutnell et al. 2015; Opanowicz et al. 2008; The International Brachypodium Initiative 2010), based on parameters from the literature and open source models.
Methods
To simulate the root and soil interactions, three open source software tools were used: 1) RootBox (Leitner et al. 2010a, 2010b; Schnepf et al. 2018), a 3D dynamic Lindenmayer System (L-systems) model of RSA, to generate the root system geometry constrained by B. distachyon root attributes reported in the literature; 2) BioCro (Jaiswal et al. 2017; Wang et al. 2015), a process-based plant biophysics model for simulating energy crops, to calculate the transpiration rates that drive the coupled root and soil water flow simulation; and 3) eSTOMP (Yabusaki et al. 2011), a variably saturated flow and multicomponent reactive transport simulator, to model the coupled process interactions given the root system geometry and boundary conditions. We first briefly describe each of the models and the parameters needed for the coupled plant root-soil system simulation for water and nutrient uptake.
Root system architecture model
Dunbabin et al. (2013) reviewed and compared six current structure-function root models that have been widely used for root modeling studies. These models are RootTyp (Pages et al. 2004), SimRoot (Lynch et al. 1997; Postma et al. 2017), ROOTMAP (Diggle 1988; Dunbabin et al. 2002), SPACSYS (Wu et al. 2007), R-SWMS (Somma et al. 1998), and RootBox (Leitner et al. 2010a; Schnepf et al. 2018). Of these six RSA models, SimRoot and RootBox are open source codes. We chose to use RootBox in this study because of its user-friendly interface and post-processing capability that made it easy to incorporate the root geometry into a soil simulation domain. RootBox is a 3D model that simulates the elongation and branching growth of individual roots according to a growth function, branching angles, and root depth defined by the user (Leitner et al. 2010a, 2010b; Schnepf et al. 2018). The L-Systems model describes the development of living things using a set of simple rules. The growth direction follows user-defined tropism. RootBox is implemented in Matlab and the code is freely available online at http://www.csc.univie.ac.at/rootbox/. The version used in this study is v6b. To be able to run this model, a Matlab license is required. However, a newer version of the model (CRootBox) written in C++ is also available for use. It is open source, more flexible and faster (Schnepf et al. 2018).
Biophysics model
BioCro is an R package that is undergoing active development (Jaiswal et al. 2017; Wang et al. 2015). It is built on a mechanistic vegetation model, WIMOVAC (Humphries and Long 1995; Song et al. 2017), which simulates and optimizes leaf-level and canopy photosynthesis, transpiration, light interception, and canopy microclimate (Miguez et al. 2012). It also simulates whole plant growth. Carbon allocation to plant structural pools is determined using dry biomass partitioning coefficients based on phenological stages (Miguez et al. 2009). Given weather condition and leaf area index, the standalone canopy function in BioCro can be used to calculate the hourly values of evapotranspiration (ET) using the Priestley-Taylor model (Priestley and Taylor 1972) option. This ET is then used to drive the root water uptake model. BioCro is freely available from https://github.com/ebimodeling/biocro. Version 0.95–8-g85cc03f was used in this study.
3D flow and reactive transport model
eSTOMP (extreme-scale Subsurface Transport Over Multiple Phases) is a highly scalable solver for simulating variably saturated subsurface flow and multicomponent, reactive transport on massively parallel processing computers (Yabusaki et al. 2011). It solves Richards’ equation for flow. The discretization of the partial differential equation is based on backward Euler in time and integral-volume, finite-difference method. Solute transport and multicomponent reactions in eSTOMP were solved using the operator-splitting approach. eSTOMP is freely available from https://github.com/pnnl/eSTOMP-WR. Version 1 was used in this study. Full documentation of the flow, transport and multicomponent reaction equations and numerical methods can be found in https://stomp.pnnl.gov/estomp_guide/eSTOMP_guide.stm.
Expressed in integral form, the mass conservation equation solved by eSTOMP is shown in Eq. (1):
where ϕ is porosity [m3 m−3], \( {\omega}_l^w \) is the mass fraction of water in the aqueous phase [kg kg−1], ρlis aqueous phase density [kg m−3], sl is aqueous phase saturation, Vn [m3] is the volume of the integration element, n is the unit surface normal vector, Γn [m2] is the surface of element, ml is the mass source/sink term [kg s−1], which is used to represent root water uptake in this study, the mass flux F is a combination of advective and diffusive components [kg m−2 s−1], calculated as the following equation:
where krl is liquid relative permeability [−], μl is kinematic viscosity [Pa s], Pl is liquid pressure [Pa], g is acceleration of gravity [m s−2], zg is unit gravitational direction vector [−], τl is liquid tortuosity [−], Mw is molecular weight of water [kg kmol−1], Ml is aqueous phase molecular weight [kg kmol−1], \( {D}_l^w \) is the diffusion coefficient of water in aqueous phase (m2 s−1), \( {\chi}_l^w \) is the mole fraction of water in aqueous phase (mol mol−1).
The conservation equation of biogeochemical species, shown in Eq. (3), equates the time rate of change of the species within a control volume with the 1) advection and diffusion/dispersion flux of species crossing the control volume surface, 2) species source/sink, and 3) the production/consumption of the species due to the biogeochemical reactions.
where Cj is the concentration of species j [kmol m−3], V is the Darcy velocity vector [m s−1], DC is the aqueous diffusion coefficient [m2 s−1], Dhl is the hydraulic dispersion tensor [m2 s−1], \( {\dot{m}}_j \) is the source rate of species j [kmol m−3 s−1], N is the number of reactions, αi, j is the stoichiometric coefficient of species j in reaction i (positive as product, negative as reactant) [−], Ri is the rate of reaction i [kmol m−3 s−1]. The biogeochemical reactions are user input based on field or laboratory evidence. They can include aqueous complexation, adsorption, ion-exchange, precipitation/dissolution, redox, acid-base reactions, and microbially mediated biological reactions.
In eSTOMP, Eq. (3) is first decomposed based on the type of biogeochemical reactions following the approach in Fang et al. (2003), resulting in a set of transport equations for primary components, transport equations for kinetic components, and mass action equations for equilibrium reactions (source/sink omitted):
in which Ec,j is the concentration of primary component j [kmol m−3], Ek,j is the concentration of the kinetic component j [kmol m−3], Nki is the number of kinetic reactions that contribute to a kinetic component, Ceq,j is the concentration of the product species for equilibrium reaction j [kmol m−3], ai is the activity of reactants for equilibrium reaction j [−], Meq,i is the number of reactants for equilibrium reaction j.
Root water uptake model
In this study, we followed the approach in Javaux et al. (2008) and Leitner et al.(2014) to simulate root water uptake, combined with soil water movement simulated by eSTOMP. For completeness, the equations solved are listed below.
Assuming the osmotic potential and the root water capacity are negligible when plants are not water stressed, the axial flow within the root is solved by discretizing the root system as a network of connected root segments:
Where Ja is the rate of axial xylem water flow rate [L3 T−1], a is the root radius [L], Ka is the axial hydraulic conductivity of the roots [L T−1], Δl is the length of root segment [L], Δz is the location difference of root segments in z direction [L], and hp is the root xylem water potential [L].
Flow between soil and root is one-dimensional radial (soil–root) flow. We made a slight modification of the equation in Javaux et al. (2008) by including the soil moisture dependence on root conductivity as reported in previous studies (Amenu and Kumar 2008) and references therein):
where Jr is the water flow rate between the soil and root [L3 T−1], \( {K}_r^{\ast } \) is the intrinsic root radial hydraulic conductivity [T−1], hs is the water potential at the soil-root interface [L], sl is the soil saturation at the soil-root interface, and sr is the outer surface area of the root segment [L2].
Javaux et al. (2008) solves Eq. (1) first and then Eqs. (8) and (9). The two steps are repeated until the solution converges. Without roots, Eq. (1) is usually solved using the standard 7-point stencil numerical approximation, where the maximum number of unknowns involved in the mass conservation of water at each grid cell is 7 – the host grid cell plus its six adjacent Cartesian neighbors (see figure in supplementary material). In this study, Eqs. (1), (8) and (9) were solved simultaneously using Newton-Raphson iterations for better computational efficiency. ml in Eq. (1) equals the negative of ρlJr in Eq. (9). For a grid cell that has root segments, the number of unknowns to be solved for mass conservation of water is 7 plus the number of root segments within that grid cell. For example, grid cell 4 in the figure shown in the supplementary material contains 3 root segments. The equation for cell 4 is a function of the unknowns at cells 1 to 7, and root segments r1, r2, and r3. For each root segment, the dependency on other unknowns can be its upstream and downstream root segments, the host grid cell which contains the root segment, and the neighboring grid cells of the host grid cell. Equation for root segment r1 depends on grid cells 1 to 7, as well as root segments r1 and r2. The equation for the grid cells without roots is only a function of the unknowns at cells 1 to 7.
The model we developed can represent dynamic growth of root systems based on root geometry calculated at different times using RootBox, prior to simulation with eSTOMP. During simulation with eSTOMP, root segments are considered inactive before they are formed and grow by activating them as they are formed, as similarly done in Daly et al. (2016). The root uptake of nutrient is assumed to be an active uptake process as opposed to a passive process where solutes are simply taken up with the porewater by the roots. The active uptake rate is represented by a Michaelis–Menten expression (Somma et al. 1998; Wu et al. 2007; Espeleta et al. 2017) in this model as shown in the following equation:
where R is the rate of nutrient uptake per root surface area [ML−2 T−1], Vmax is the maximum uptake rate [ML−2 T−1], [C is the soil solute concentration adjacent to the root [ML−3], and Km is the substrate affinity constant [ML−3]. The uptake is represented as a sink term (removing nutrients from the soil) in the reactive transport equation of eSTOMP. We assume Vmax and Km are the same for a specific nutrient, but they can be specified by root segment if necessary. The uptake rate changes spatially along the roots as the concentration is a function of soil moisture, transport, nutrient uptake, and multicomponent reactions adjacent to each root segment.
Simulation domain
The simulation domain is 36 cm × 36 cm × 48 cm. Nominal grid resolution for the base case simulation is 1 cm in each direction. The van Genuchten function soil water retention parameters are from a simulation scenario presented in Javaux et al. (2008): α = 0.036 cm−1, n = 1.56, soil porosity = 0.43, and saturated hydraulic conductivity = 250 cm d−1. We use radial and axial root conductivities similar to those used by Javaux et al. (2008) and Amenu and Kumar (2008), which are 2.0 × 10−8 s−1 and 3.97 × 10−3 mm s−1, respectively.
Initial and boundary conditions
For the soil system, a constant water potential of −3.0 m is applied to the west and east sides of the model domain, and the soil water is initially in equilibrium with the boundary conditions. These boundary conditions act to maintain bulk water content in the system by replenishing water from the boundaries as water is taken up by the roots. Note that these are simplified boundary conditions as in reality water recharge typically comes from precipitation or irrigation. No-flux boundary conditions are applied elsewhere. Assuming a preformed root system, a 24 h time-varying transpiration cycle calculated using BioCro is repeated at the root collar during the simulation period (14 days).
Competitive cation exchange
Using a simple 1D model given prescribed water potential at soil-root boundary, Espeleta et al. (2017) found that competitive cation exchange enabled NH4+ and K+ from soil to be desorbed from soil exchange sites and made available for plant use when low-demand cations such as Ca2+, Mg2+, and Na+ accumulated near roots during water uptake. To simulate competitive ion exchange in 3D, cation exchange reactions (Table 1) and root nutrient uptake were included in our model. The selectivity constants in Table 1 were derived from literature reported values (Liu et al. 2009; Yan et al. 2018). The cation exchange capacity and bulk density of the soil are 20 meq 100 g−1 and 1.3 g cm−3, respectively. The clay content of the soil is 9%. Initial aqueous concentrations (Table 2) are in equilibrium with the exchange sites. The Gaines-Thomas cation exchange modeling convention (Gaines and Thomas 1953), i.e., activities of cations on exchange sites are expressed as cation equivalent fraction, is used in our model. As there are no kinetic reactions in this problem specification, eSTOMP only needs to solve the transport equation for primary components (Eq. 5) and mass action equations (Eq. 7). In the general case with mixed kinetic and equilibrium reactions, eSTOMP partitions the kinetic species for additional transport calculations. The aqueous diffusion coefficient is 1.6 × 10−9 m2 s−1. The longitudinal and transverse dispersivities are 0.1 cm and 0.01 cm, respectively. The other input parameters are taken from Espeleta et al. (2017) and shown in Table 2 for completeness. Cl− is a conservative tracer (assumed to not be taken up by the root and not be reactive) in this simulation, included for charge balance. Charge-equivalents of H+ are released through the root-soil interface to balance the uptake of cations. While the simulator can accommodate large complex biogeochemical reaction networks (Yabusaki et al. 2017), a simple abbreviated set of reactions (Table 1) is used for this demonstration. Cauchy boundary conditions are set at the east and west boundary of the simulation domain. Concentrations flowing in are the same as the initial solute concentration in the soil. The flow condition is the same as described in the section above. The 24-h boundary transpiration conditions are repeated every day for the length (14 d) of the simulation. The principal flow dynamics are driven by the diel root water uptake.
RSA
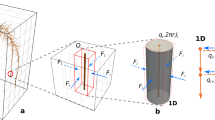
RSA of B. distachyon consists of three root types: primary (seminal), coleoptile node (emerging from a node above seed and below the soil surface), and leaf node roots (or shoot-borne) roots. B. distachyon root length reported in the literature showed that 6 weeks after planting, leaf node axile roots (LNRs) contribute more than 50% of the total root length (Donn et al. 2017; Watt et al. 2009). Taking this into account, we generated the root system geometry for B. distachyon in a container of 36 cm × 36 cm × 48 cm size in a planting time-frame of 6 weeks (Fig. 1a). The geometric complexity of the simulated root system resembles B. distachyon RSA grown in growth chamber under controlled condition in identical pot size (Fig. 1b). The spatial resolution along the root axis was 0.5 cm. The diameter of B. distachyon roots ranged from 47 to 500 μm (Donn et al. 2017; Watt et al. 2009). We set a uniform root diameter of 250 μm in this study. The resulting RSA consisted of 27 individual root branches comprising 2194 root segments, a root system depth of 45.8 cm, and a total root length of 1097 cm at day 42, the end of the simulation. The primary axile root (RP) length is 100 cm, and the coleoptile node axile root (CNR) length is 186 cm. The simulated LNR contributes 74% of total root length. A root fraction profile created based on the number of root segments in each grid cell layer shows that 90% of the root is within the top 24 cm. Overall, this simulated root system shows the complex root-bound geometries that can be handled by our model.
Transpiration
Donn et al. (2017) studied the transpiration efficiency of seven B. distachyon accessions grown in field soils. They measured photosynthesis and transpiration rates approximately seven weeks after planting using a LI-6400 Portable Photosynthesis System (LI-COR). LI-COR is a gas exchange analyzer widely used to quantify CO2 and water fluxes across a leaf surface. It has two absolute CO2 and two absolute H2O non-dispersive infrared analyzers in the sensor head allowing a real time measurement of changes in the leaf dynamics. In the study by Donn et al. (2017), the chamber temperature was set to 25 °C and light was set at 1000 μmol of photons m−2 s−1 for the measurements. The measurements of six replicates for each B. distachyon accession showed the transpiration rate ranged from 0.27 to 8.30 mmol/m2/s.
Using the default C3 grass parameterization in BioCro, the example weather data “doy124” from the BioCro package, and an assumed leaf area index of 1.36, a transpiration rate (Fig. 2) comparable to those measured by Donn et al. (2017) was generated. This rate was converted to volume rate of water using canopy area (we assume it’s the same as the container area) in our simulation.
Results
Flow results
Using the above generated RSA and the 24 h cycle of transpiration rate dynamics as a repeating boundary condition for the 14-day simulation period of coupled flow and reactive transport, it can be seen that at 15 h on day 14, the highest pressure head decrease at the center of the cross section taken at 6 cm belowground is 0.57 m lower than the background pressure head (Fig. 3). Changes decrease away from the center of the root mass, which suggests it is beneficial to have a larger branched RSA for the water needs of plants because the distribution of transpiration-driven flow over more root surface area would result in less extreme desaturation along the soil-root interface.
Soil saturation changes due to diel root water uptake. At 2 cm belowground, the contour of saturation during the middle of the day (Fig. 4a) and night (Fig. 4b) of day 14 shows that the impact of root water uptake is contained within 4 cm of the roots. The size of the impact zone is sensitive to grid resolution. When the grid size is decreased by half, more localized saturation distribution at the same location is simulated (Fig. 4c, d). During the dark periods, the rhizosphere water is replenished by water from the surrounding soil, decreasing the saturation gradient from the wall of the container to the rhizosphere (Fig. 4b).
Summing the soil to root flow rate for each root segment in a soil layer, the total flow rate between the soil and root in each layer can be obtained and is shown in Fig. 5. The rate is positive when the root takes in water and negative when water is released to the soil from the root. It can be seen that during the dark, deep roots take in water from deeper soil layers and release water into the upper soil layer. The flux is larger at the top 10 cm due to higher root fractions. Compared to the simple treatment of transpiration as sink terms in soil layers, this model can be better constrained by measurable parameters, such as xylem potential, root hydraulic conductivity etc.
Cation exchange results
Initially, the order of abundance on the exchange sites is: Ca2+ > Mg2+ > Na+ > NH4+ > K+ > H+. Sorbed calcium, CaX2, is the dominant species on the exchange sites. Due to the inter-root competition for water which results in spatially variable soil moisture distribution in the soil, the solute concentration isosurface in the solution is non-uniformly distributed in space (Fig. 6).
During the day, root water uptake desaturates soil pores, which results in the buildup of solution concentrations near the roots (Fig. 7a). Percentage change of Cl− concentration between the night and the middle of the day (Fig. 7b) shows that aqueous concentrations are redistributed at night due, in part, to re-saturation in the rhizosphere, low transpiration and diffusion. Concentration at night is slightly higher (positive percentage change) away from the root as the saturation is lower in those areas compared to the day time (Fig. 4a, b). Saturation is highest at the end of the dark period each day adjacent to the roots, causing small dilution of aqueous Cl− concentration. Because we assume only active uptake of ions, no ions are explicitly taken up in root water. The mixing is not strong enough to dilute the concentration, resulting in much higher concentration compared to the background solution.
Two horizontal planes were selected to look at the aqueous and sorbed exchanger concentrations as factors of their initial states (states at the beginning of the simulation). One at 2 cm belowground where there is one root segment centered at x = 0 cm (Fig. 8a, b), and the other is at 8 cm belowground, which is affected by multiple root segments (Fig. 8c, d). During the daytime, movement of water toward individual roots due to root water uptake desaturates soil pore water, building up solution concentrations near the roots. Compared to Cl−, both locations show that 1) Ca2+ and Mg2+ are desorbed from the exchange site, while Na+, K+ and NH4+ are sorbed to the site due to the increase of solute concentrations caused by the root water uptake; 2) concentration of monovalent ions decreased in the solution due to root uptake and sorption. This finding differs from the 1D model in Espeleta et al. (2017) where NH4+ and K+ from soil are desorbed due to the buildup of low demand ions in solution. This is due to the parameters used in the different simulations. To test parameter sensitivity, we increased all of the ion uptake rates by a factor of 10 and found that when the uptake rate is faster than the accumulation rate due to root zone desaturation, the buildup of less preferred Na+ causes desorption of NH4+ from the exchange site and increase of NH4+ in the solution.
Aqueous (a) and exchanger species (b) concentration as a factor of initial condition for Y = 18 cm and Z = 2 cm belowground during the middle of the day on day 14, and aqueous (c) and exchanger species (d) concentration as a factor of initial condition at t = 336 h for Y = 18 cm and Z = 8 cm belowground during the middle of day on day 14. The peak concentration for (a) and (b) is centered around the root
The bulk concentration behavior away from the center of root mass is similar at deep and shallow locations with minimal variation. Conversely, the local nutrient concentrations near a root segment can be dynamic and spatially variable due to root water uptake and release, nutrient uptake and buildup of ions, ion exchange, and interactions between nearby roots.
Discussion
Taking advantage of open source software for plant biophysics, root growth, variably saturated flow and multicomponent reactive transport, we developed a simulation capability to address plant-soil interactions via integration of diurnal transpiration, water and ion uptake and release from 3-D root system architecture, soil-root xylem flow, and coupled hydrobiogeochemical processes in soil. The parallel processing formulation of the simulator exploits high performance computing to enable tractable run times for simulations of highly-resolved root system architecture and soil hydrobiogeochemical processes. Eight processor cores were used for each of the coupled flow and reactive transport simulations of our hypothetical problem. All were completed in less than 1 h of wall clock time. This efficient capability that considers two-way hydrologic coupling between the soil and RSA and multicomponent reactive transport in the soil for nutrient uptake has not previously been reported. It can better represent the water and nutrient status in the soil and hence their availability for plant use through interactions of physical and biogeochemical processes between plant roots and soil. Though not shown in our hypothetical application, it is convenient for users to develop models to explore soil biogeochemical processes including C, N, P etc. in the rhizosphere as these biogeochemical reactions and rates can be provided as user input.
Demonstrated on B. distachyon, we were able to simulate the complex moisture and concentration distribution near the roots due to inter-root competition and multicomponent competitive ion exchange. The model simulated upward hydraulic lift during the night, i.e., deep roots take in water from deeper soil layers and release water into the upper drier soil layers which can then be used by shallow roots in the daytime, and enhance nutrient availability to plants (Cardon et al. 2013). This hydraulic redistribution has important consequences for stomatal opening, transpiration, and overall carbon gain (Cardon et al. 2013). While this is a hypothetical simulation, it is worth noting that root pore water uptake results in generally higher concentrations for all cations despite the preferential uptake of K+ and NH4+. This leads to the seemingly paradoxical increase in lower affinity cations on the exchange sites. This behavior actually follows the ratio law of Schofield (Schofield 1947), i.e., increasing preference for the lower valent ion as the solution concentrations of all cations increase. Only when our model parameters were varied, were we able to identify the conditions for increased availability of high-demand cations due to competitive ion exchange as reported in the literature. This underscores the complexity of coupled hydrobiogeochemical processes in the rhizosphere. Our demonstration also implies that the assumption of steady state water conditions (i.e., desaturation not being represented) in the soil will result in the underestimation of nutrient uptake by plants under nutrient-deficient conditions.
Admittedly, the model could be made more realistic with the addition of microbiological reactions, more complex soil chemistry, rhizodeposition, spatial variation of root function, xylem vulnerability, and soil water feedback to canopy transpiration under stress conditions. As noted by Carminati et al. (2016), these types of root water and nutrient uptake models require many parameters that are not easily measurable. However, with advances in imaging technologies and comprehensive field or laboratory studies, the framework of this tool makes it possible to explore different conceptual models to improve the understanding of the biogeochemical and physical environment around the roots and the flow of carbon and nutrients in ecosystems.
Conclusion
A high performance computing rhizosphere modeling capability has been developed using open source software components (Fig. 9). The capability explicitly represents individual roots in a 3D root system. Water potential in the root xylem network and soil system is fully coupled and solved simultaneously. Root water uptake and release leads to spatiotemporal variations in saturation and water flow. Concentration-dependent nutrient uptake by the roots is mediated by desaturation, transport, and multicomponent reactions. The schematics of the coupling and the simple reactions we demonstrated are shown in Fig. 9.
The schematics of process coupling and the simple reactions (cation exchange between the plant root, soil solution and exchangeable sites on the right was modified from OMAFRA Publication 811 ( 2006))
The technology was demonstrated on a soil-root system based on B. distachyon using parameters and conditions reported in the literature. A root system architecture was generated using a 3D dynamic L-systems model of root architecture. Transpiration at the root collar was generated using plant biophysics process modeling, eSTOMP (Yabusaki et al. 2011) was modified to simulate the coupled soil-root xylem flow, nutrient uptake, and competitive ion exchange that resulted in spatiotemporal soil water potential and saturation, and nutrient concentrations. The simulator developed in this study will be made publicly available. The principal conclusion is that rhizosphere nutrient concentrations are the result of multiple interacting processes encompassing plant biophysics, root system architecture, soil-root xylem flow, nutrient uptake, and multicomponent reactive transport. While an actual experiment was not modeled, insights were gained into the interplay of processes and the sensitivity of behaviors to grid resolutions, root attributes, geochemistry, and soil properties. In general, this underscores the view of Hinsinger et al. (2008), that nutrient dynamics should be tied to water dynamics.
References
Amenu GG, Kumar P (2008) A model for hydraulic redistribution incorporating coupled soil-root moisture transport. Hydrol Earth Syst Sci 12:55–74
Brutnell TP, Bennetzen JL, Vogel JP (2015) Brachypodium distachyon and Setaria viridis: model genetic systems for the grasses. Annu Rev Plant Biol 66:465–485
Cardon ZG, Stark JM, Herron PM, Rasmussen JA (2013) Sagebrush carrying out hydraulic lift enhances surface soil nitrogen cycling and nitrogen uptake into inflorescences. Proc Natl Acad Sci U S A 110:18988–18993
Carminati A, Zarebanadkouki M, Kroener E, Ahmed MA, and Holz M (2016) Biophysical rhizosphere processes affecting root water uptake, Ann Bot-London, 118(4), 561-571
Christoffersen BO, Gloor M, Fauset S, Fyllas NM, Galbraith DR, Baker TR, Kruijt B, Rowland L, Fisher RA, Binks OJ, Sevanto S, Xu CG, Jansen S, Choat B, Mencuccini M, McDowell NG, Meir P (2016) Linking hydraulic traits to tropical forest function in a size-structured and trait-driven model (TFS v.1-hydro). Geosci Model Dev 9:4227–4255
Chung JB, Zasoski RJ (1994) Ammonium-potassium and ammonium-calcium exchange equilibria in bulk and rhizosphere soil. Soil Sci Soc Am J 58:1368–1375
Clausnitzer V, Hopmans JW (1994) Simultaneous modeling of transient 3-dimensional root-growth and soil-water flow. Plant Soil 164:299–314
Daly KR, Keyes SD, Masum S, Roose T (2016) Image-based modelling of nutrient movement in and around the rhizosphere. J Exp Bot 67:1059–1070
Diggle AJ (1988) Rootmap - a aodel in 3-dimensional coordinates of the growth and structure of fibrous root systems. Plant Soil 105:169–178
Donn S, Kawasaki A, Delroy B, Chochois V, Watt M, Powell JR (2017) Root type is not an important driver of mycorrhizal colonisation in &ITBrachypodium distachyon&IT. Pedobiologia 65:5–15
Doussan C, Pierret A, Garrigues E, Pages L (2006) Water uptake by plant roots: II - modelling of water transfer in the soil root-system with explicit account of flow within the root system - comparison with experiments. Plant Soil 283:99–117
Dunbabin VM, Diggle AJ, Rengel Z, van Hugten R (2002) Modelling the interactions between water and nutrient uptake and root growth. Plant Soil 239:19–38
Dunbabin V, Rengel Z, Diggle AJ (2004) Simulating form and function of root systems: efficiency of nitrate uptake is dependent on root system architecture and the spatial and temporal variability of nitrate supply. Funct Ecol 18:204–211
Dunbabin VM, McDermott S, Bengough AG (2006) Upscaling from rhizosphere to whole root system: modelling the effects of phospholipid surfactants on water and nutrient uptake. Plant Soil 283:57–72
Dunbabin VM, Postma JA, Schnepf A, Pages L, Javaux M, Wu LH, Leitner D, Chen YL, Rengel Z, Diggle AJ (2013) Modelling root-soil interactions using three-dimensional models of root growth, architecture and function. Plant Soil 372:93–124
Espeleta JF, Cardon ZG, Mayer KU, Neumann RB (2017) Diel plant water use and competitive soil cation exchange interact to enhance NH4 (+) and K+ availability in the rhizosphere. Plant Soil 414:33–51
Fang YL, Yeh GT, Burgos WD (2003) A general paradigm to model reaction-based biogeochemical processes in batch systems. Water Resour Res 39
Finzi AC, Abramoff RZ, Spiller KS, Brzostek ER, Darby BA, Kramer MA, Phillips RP (2015) Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob Chang Biol 21:2082–2094
Gaines GL, Thomas HC (1953) Adsorption studies on clay minerals .2. A formulation of the thermodynamics of exchange adsorption. J Chem Phys 21:714–718
Garcia-Tejera O, Lopez-Bernal A, Testi L, Villalobos FJ (2017) A soil-plant-atmosphere continuum (SPAC) model for simulating tree transpiration with a soil multi-compartment solution. Plant Soil 412:215–233
Garrigues E, Doussan C, Pierret A (2006) Water uptake by plant roots: I - formation and propagation of a water extraction front in mature root systems as evidenced by 2D light transmission imaging. Plant Soil 283:83–98
Gérard F, Blitz-Frayret C, Hinsinger P, Pagès L (2017) Modelling the interactions between root system architecture, root functions and reactive transport processes in soil. Plant Soil 413:161–180
Helliwell JR, Sturrock CJ, Mairhofer S, Craigon J, Ashton RW, Miller AJ, Whalley WR, Mooney SJ (2017) The emergent rhizosphere: imaging the development of the porous architecture at the root-soil interface. ScientificReports, 7:Artn 14875
Hinsinger P, Bravin MN, Devau N, Gerard F, Le Cadre E, Jaillard B (2008) Soil-root-microbe interactions in the rhizosphere - a key to understanding and predicting nutrient bioavailability to plants. J. Soil Sci. Plant Nutr 8, 39-47
Humphries SW, Long SP (1995) WIMOVAC - a software package for modeling the dynamics of plant leaf and canopy hotosynthesis. Comput Appl Biosci 11:361–371
Jaiswal D, De Souza AP, Larsen S, LeBauer DS, Miguez FE, Sparovek G, Bollero G, Buckeridge MS, Long SP (2017) Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nat Clim Chang 7:788–792
Javaux M, Schroder T, Vanderborght J, Vereecken H (2008) Use of a three-dimensional detailed modeling approach for predicting root water uptake. Vadose Zone J 7:1079–1088
Leitner D, Klepsch S, Bodner G, Schnepf A (2010a) A dynamic root system growth model based on L-systems. Plant Soil 332:177–192
Leitner D, Klepsch S, Kniess A, Schnepf A (2010b) The algorithmic beauty of plant roots - an L-system model for dynamic root growth simulation. Math Comput Model Dyn Syst 16:575–587
Leitner D, Schnepf A, Klepsch S, Roose T (2010c) Comparison of nutrient uptake between 3-dimensional simulation and an averaged root system model. Plant Biosystems 144:5
Leitner D, Meunier F, Bodner G, Javaux M, Schnepf A (2014) Impact of contrasted maize root traits at flowering on water stress tolerance - a simulation study. Field Crop Res 165:125–137
Liu CX, Shi ZQ, Zachara JM (2009) Kinetics of uranium(VI) desorption from contaminated sediments: effect of geochemical conditions and model valuation. Environ Sci Technol 43:6560–6566
Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287
Lynch JP, Nielsen KL, Davis RD, Jablokow AG (1997) SimRoot: modelling and visualization of root systems. Plant Soil 188:139–151
Manoli G, Bonetti S, Domec JC, Putti M, Katul G, Marani M (2014) Tree root systems competing for soil moisture in a 3D soil-plant model. Adv Water Resour 66:32–42
Manoli G, Huang CW, Bonetti S, Domec JC, Marani M, Katul G (2017) Competition for light and water in a coupled soil-plant system. Adv Water Resour 108:216–230
Matamala R, Stover DB (2013) Introduction to a virtual special issue: modeling the hidden half - the root of our problem. New Phytol 200:939–942
Miguez FE, Zhu XG, Humphries S, Bollero GA, Long SP (2009) A semimechanistic model predicting the growth and production of the bioenergy crop Miscanthus x giganteus: description, parameterization and validation. Glob Change Biol Bioenergy 1:282–296
Miguez FE, Maughan M, Bollero GA, Long SP (2012) Modeling spatial and dynamic variation in growth, yield, and yield stability of the bioenergy crops Miscanthus x giganteus and Panicum virgatum across the conterminous United States. Glob Change Biol Bioenergy 4:509–520
Mollier A, De Willigen P, Heinen M, Morel C, Schneider A, Pellerin S (2008) A two-dimensional simulation model of phosphorus uptake including crop growth and P-response. Ecol Model 210:453–464
Nietfeld H, Prenzel J (2015) Modeling the reactive ion dynamics in the rhizosphere of tree roots growing in acid soils. I. Rhizospheric distribution patterns and root uptake of M-b cations as affected by root-induced pH and Al dynamics. Ecol Model 307:48–65
OMAFRA (2006) Soil fertility handbook publication 611. Ontario Ministry of Agriculture, Food and Rural Affairs, Toronto
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177
Pages L, Vercambre G, Drouet JL, Lecompte F, Collet C, Le Bot J (2004) Root Typ: a generic model to depict and analyse the root system architecture. Plant Soil 258:103–119
Postma JA, Kuppe C, Owen MR, Mellor N, Griffiths M, Bennett MJ, Lynch JP, Watt M (2017) OPENSIMROOT: widening the scope and application of root architectural models. New Phytol 215:1274–1286
Priestley CHB, Taylor RJ (1972) Assessment of surface heat-flux and evaporation using large-scale parameters. Mon Weather Rev 100:81–8+
Schnepf A, Leitner D, Landl M, Lobet G, Mai TH, Morandage S, Sheng C, Zorner M, Vanderborght J, Vereecken H (2018) CRootBox: a structural-functional modelling framework for root systems. Ann Bot 121:1033–1053
Schofield RK (1947) A ratio law governing the equilibrium of cations in the soil solution Proc 2nd Int Congr Pure Appl Chem 3: 257-261
Simunek J, Hopmans JW (2009) Modeling compensated root water and nutrient uptake. Ecol Model 220:505–521
Somma F, Hopmans JW, Clausnitzer V (1998) Transient three-dimensional modeling of soil water and solute transport with simultaneous root growth, root water and nutrient uptake. Plant Soil 202:281–293
Song QF, Chen DR, Long SP, Zhu XG (2017) A user-friendly means to scale from the biochemistry of photosynthesis to whole crop canopies and production in time and space - development of Java WIMOVAC. Plant Cell Environ 40:51–55
Sperry JS, Adler FR, Campbell GS, Comstock JP (1998) Limitation of plant water use by rhizosphere and xylem conductance: results from a model. Plant Cell Environ 21:347–359
The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763
Tournier PH, Hecht F, Comte M (2015) Finite element model of soil water and nutrient transport with root uptake: explicit geometry and unstructured adaptive meshing. Transp Porous Media 106:487–504
Vereecken H, Schnepf A, Hopmans JW, Javaux M, Or D, Roose DOT, Vanderborght J, Young MH, Amelung W, Aitkenhead M, Allison SD, Assouline S, Baveye P, Berli M, Bruggemann N, Finke P, Flury M, Gaiser T, Govers G, Ghezzehei T, Hallett P, Franssen HJH, Heppell J, Horn R, Huisman JA, Jacques D, Jonard F, Kollet S, Lafolie F, Lamorski K, Leitner D, McBratney A, Minasny B, Montzka C, Nowak W, Pachepsky Y, Padarian J, Romano N, Roth K, Rothfuss Y, Rowe EC, Schwen A, Simunek J, Tiktak A, Van Dam J, van der Zee SEATM, Vogel HJ, Vrugt JA, Wohling T, Young IM (2016) Modeling soil processes: review, key challenges, and new perspectives. Vadose Zone J 15
Wang D, Jaiswal D, Lebauer DS, Wertin TM, Bobbero GA, Leakey ADB, Long SP (2015) A physiological and biophysical model of coppice willow (Salix spp.) production yields for the contiguous USA in current and future climate scenarios. Plant Cell Environ 38:1850–1865
Watt M, Schneebeli K, Dong P, Wilson IW (2009) The shoot and root growth of Brachypodium and its potential as a model for wheat and other cereal crops. Funct Plant Biol 36:960–969
Wu L, McGechan MB, McRoberts N, Baddeley JA, Watson CA (2007) SPACSYS: integration of a 3D root architecture component to carbon, nitrogen and water cycling-model description. Ecol Model 200:343–359
Yabusaki SB, Fang YL, Williams KH, Murray CJ, Ward AL, Dayvault RD, Waichler SR, Newcomer DR, Spane FA, Long PE (2011) Variably saturated flow and multicomponent biogeochemical reactive transport modeling of a uranium bioremediation field experiment. J Contam Hydrol 126:271–290
Yabusaki SB, Wilkins MJ, Fang Y, Williams KH, Arora B, Bargar J, Beller HR, Bouskill NJ, Brodie EL, Christensen JN, Conrad ME, Danczak RE, King E, Soltanian MR, Spycher NF, Steefel CI, Tokunaga TK, Versteeg R, Waichler SR, Wainwright HM (2017) Water table dynamics and biogeochemical cycling in a shallow, variably-saturated floodplain. Environ Sci Technol 51:3307–3317
Yan A, Liu C, Liu Y, Xu F (2018) Effect of ion exchange on the rate of aerobic microbial oxidation of ammonium in hyporheic zone sediments. Environ Sci Pollut Res
York LM, Carminati A, Mooney SJ, Ritz K, Bennett MJ (2016) The holistic rhizosphere: integrating zones, processes, and semantics in the soil influenced by roots. J Exp Bot 67:3629–3643
Zhu XG, Lynch JP, LeBauer DS, Millar AJ, Stitt M, Long SP (2016) Plants in silico: why, why now and what?An integrative platform for plant systems biology research. Plant Cell Environ 39:1049–1057
Acknowledgments
This work was funded in part by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research under contract DE-AC05-76RL01830 with Pacific Northwest National Laboratory through the iPASS and PREMIS Initiatives. A portion of this research was performed using Institutional Computing at PNNL. PNNL is operated for the U.S. Department of Energy by Battelle Memorial Institute under contract DE-AC05-76RL01830. We thank Dr. Suzanne Donn at Western Sydney University for providing the experiment data. We also thank the anonymous reviewers whose constructive comments have greatly improved our manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Andrea Schnepf.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 85 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fang, Y., Yabusaki, S.B., Ahkami, A.H. et al. An efficient three-dimensional rhizosphere modeling capability to study the effect of root system architecture on soil water and reactive transport. Plant Soil 441, 33–48 (2019). https://doi.org/10.1007/s11104-019-04068-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04068-z