Abstract
Aims
To understand effects of tissue type, growth stage and soil fertilisers on bacterial endophyte communities of winter wheat (Triticum aestivum cv. Hereward).
Methods
Endophytes were isolated from wheat grown under six fertiliser conditions in the long term Broadbalk Experiment at Rothamsted Research, UK. Samples were taken in May and July from root and leaf tissues.
Results
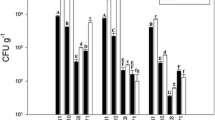
Root and leaf communities differed in abundance and composition of endophytes. Endophytes were most abundant in roots and the Proteobacteria were most prevalent. In contrast, Firmicutes and Actinobacteria, the Gram positive phyla, were most prevalent in the leaves. Both fertiliser treatment and sample time influenced abundance and relative proportions of each phylum and genus in the endosphere. A higher density of endophytes was found in the Nil input treatment plants.
Conclusions
Robust isolation techniques and stringent controls are critical for accurate recovery of endophytes. The plant tissue type, plant growth stage, and soil fertiliser treatment all contribute to the composition of the endophytic bacterial community in wheat. These results should help facilitate targeted development of endophytes for beneficial applications in agriculture.









Similar content being viewed by others
References
Barroso JM (2011) Regulations: Commission implementing regulation (EC) No 1107/2009 of the the European Parliament and of the council as regards the list of approved active substances. Official Journal of the European Union 153.
Bodenhausen N, Horton MW, Bergelson J (2013) Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8, e56329. doi:10.1371/journal.pone.0056329
Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi:10.1038/nature11336
Chelius MK, Triplett EW (2001) The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol 41:252–263. doi:10.1007/s002480000087
Clark IM, Buchkina N, Jhurreea D, Goulding KW, Hirsch PR (2012) Impacts of nitrogen application rates on the activity and diversity of denitrifying bacteria in the Broadbalk Wheat Experiment. Philos Trans R Soc Lond B Biol Sci 367:1235–1244. doi:10.1098/rstb.2011.0314
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/nar/gkn879
de Santi Ferrara FI, Oliveira ZM, Gonzales HHS, Floh EIS, Barbosa HR (2012) Endophytic and rhizopheric enterobacteria isolated from sugar cane have different potentials for producing plant growth-promoting sunstances. Plant Soil 353:409–417. doi:10.1007/s11104-011-1042-1
Digby PGN, Kempton RA (1987) Multivariate analysis of ecological communities. Chapmann and Hall, London
Ferrando L, Fernandez Manay J, Fernandez Scavino A (2012) Molecular and culture-dependent analyses revealed similarities in the endophytic bacterial community composition of leaves from three rice (Oryza sativa) varieties. FEMS Microbiol Ecol 80:696–708. doi:10.1111/j.1574-6941.2012.01339.x
Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B (2007) Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci 26:227–242. doi:10.1080/07352680701572966
Gupta M, Kiran S, Gulati A, Singh B, Tewari R (2012) Isolation and identification of phosphate solubilizing bacteria able to enhance the growth and aloin-A biosynthesis of Aloe barbadensis Miller. Microbiol Res 167:358–363. doi:10.1016/j.micres.2012.02.004
Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43:895–914
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Paleontol Electron 4:9
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471. doi:10.1016/j.tim.2008.07.008
Hardoim PR, Andreote FD, Reinhold-Hurek B, Sessitsch A, van Overbeek LS, van Elsas JD (2011) Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol Ecol 77:154–164. doi:10.1111/j.1574-6941.2011.01092.x
Johnston-Monje D, Raizada MN (2011) Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6, e20396. doi:10.1371/journal.pone.0020396
Kembel SW, Wu M, Eisen JA, Green JL (2012) Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol 8(10):e1002743. doi:10.1371/journal.pcbi.1002743.g001
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41(1):e1. doi:10.1093/nar/gks808
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40(9):2407–2415. doi:10.1016/j.soilbio.2008.05.021
Lopez Brucio J, Nieto-Jacobo M, Ramirez-Rodriguez V, Herrera-Estrella L (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160:1–13
Lopez BR, Tinoco-Ojanguren C, Bacilio M, Mendoza A, Bashan Y (2012) Endophytic bacteria of the rock-dwelling cactus Mammillaria fraileana affect plant growth and mobilization of elements from rocks. Environ Exp Bot 81:26–36. doi:10.1016/j.envexpbot.2012.02.014
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi:10.1038/nature11237
Mano H, Tanaka F, Nakamura C, Kaga H, Morisaki H (2007) Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ 22:175–185
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795
Marschner P, Kandeler E, Marschner B (2003) Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol Biochem 35:453–461. doi:10.1016/s0038-0717(02)00297-3
Monteiro RA, Balsanelli E, Wassem R, Marin AM, Brusamarello-Santos LCC, Schmidt MA, Tadra-Sfeir MZ, Pankievicz VCS, Cruz LM, Chubatsu LS, Pedrosa FO, Souza EM (2012) Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant Soil 356:175–196. doi:10.1007/s11104-012-1125-7
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
OECD/FAO (2011) OECD-FAO Agricultural Outlook 2011–2020. OECD Publishing and FAO, Rome, Italy
Olesen JE, Trnka M, Kersebaum KC, Skjelvåg AO, Seguin B, Peltonen-Sainio P, Rossi F, Kozyra J, Micale F (2011) Impacts and adaptation of European crop production systems to climate change. Eur J Agron 34:96–112. doi:10.1016/j.eja.2010.11.003
Osborn AM, Moore ER, Timmis KN (2000) An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol 2:39–50
Pavlo A, Leonid O, Iryna Z, Natalia K, Maria PA (2011) Endophytic bacteria enhancing growth and disease resistance of potato (Solanum tuberosum L.). Biol Control 56:43–49. doi:10.1016/j.biocontrol.2010.09.014
Pineda A, Zheng SJ, van Loon JJ, Dicke M (2012) Rhizobacteria modify plant-aphid interactions: a case of induced systemic susceptibility. Plant Biol 14:83–90. doi:10.1111/j.1438-8677.2011.00549.x
Ramesh R, Joshi AA, Ghanekar MP (2009) Pseudomonads: major antagonistic endophytic bacteria to suppress bacterial wilt pathogen, Ralstonia solanacearum in the eggplant (Solanum melongena L.). World J Microbiol Biotechnol 25:47–55
Redford AJ, Fierer N (2009) Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58:189–198. doi:10.1007/s00248-009-9495-y
Rees RM, Baddeley JA, Bhogal A, Ball BC, Chadwick DR, Macleod M, Lilly A, Pappa VA, Thorman RE, Watson CA, Williams JR (2013) Nitrous oxide mitigation in UK agriculture. Soil Sci Plant Nutr 59:3–15. doi:10.1080/00380768.2012.733869
Reinhold-Hurek B, Hurek T (2011) Living inside plants: bacterial endophytes. Curr Opin Plant Biol 14:435–443. doi:10.1016/j.pbi.2011.04.004
Ren J-H, Li H, Wang Y-F, Ye J-R, Yan A-Q, Wu X-Q (2013) Biocontrol potential of an endophytic Bacillus pumilus JK-SX001 against poplar canker. Biol Control 67:421–430. doi:10.1016/j.biocontrol.2013.09.012
Rothamsted (2006) Guide to the classical and other long-term experiments, datasets and sample archive. Lawes Agricultural Trust Co. Ltd, Harpenden, UK
Seghers D, Wittebolle L, Top EM, Verstraete W, Siciliano SD (2004) Impact of agricultural practices on the Zea mays L. endophytic community. Appl Environ Microbiol 70:1475–1482. doi:10.1128/aem.70.3.1475-1482.2004
Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomyces-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 39:23–32
Thaweenut N, Hachisuka Y, Ando S, Yanagisawa S, Yoneyama T (2010) Two seasons’ study on nifH gene expression and nitrogen fixation by diazotrophic endophytes in sugarcane (Saccharum spp. hybrids): expression of nifH genes similar to those of rhizobia. Plant Soil 338:435–449. doi:10.1007/s11104-010-0557-1
Thrall PH, Hochberg ME, Burdon JJ, Bever JD (2006) Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol 22(3):120–126
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant A, Poole PS (2013) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7(12):2248–2258. doi:10.1038/ismej.2013.119
van Grinsven HJM, Spiertz JHJ, Westhoek HJ, Bouwman AF, Erisman JW (2013) Nitrogen use and food production in European regions from a global perspective. The Journal of Agricultural Science Nitrogen Workshop Special Issue Paper: 1–11. doi: 10.1017/s0021859613000853
Watts CW, Clark LJ, Poulton PR, Powlson DS, Whitmore AP (2006) The role of clay, organic carbon and long-term management on mouldboard plough draught measured on the Broadbalk wheat experiment at Rothamsted. Soil Use Manag 22:334–341. doi:10.1111/j.1475-2743.2006.00054.x
Wilson D (1995) Endophyte: the evolution of a term, and clarification of its use and definition. Oikos 73:274–276
Zhalnina K, de Quadros PD, Gano KA, Davis-Richardson A, Fagen JR, Brown CT, Giongo A, Drew JC, Sayavedra-Soto LA, Arp DJ, Camargo FA, Daroub SH, Clark IM, McGrath SP, Hirsch PR, Triplett EW (2013) Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front Microbiol 4:104. doi:10.3389/fmicb.2013.00104
Acknowledgments
We are grateful to the Biotechnology and Biological Sciences Research Council (BBSRC) and to Novozymes for providing the funding for a PhD studentship for R. Robinson. We would particularly like to thank Dr Stephen Powers (Rothamsted Research) for his help and advice with statistical analysis and Dr Ben Raymond (Imperial College) for statistical advice and recommendations.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Andrea Campisano.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Robinson, R.J., Fraaije, B.A., Clark, I.M. et al. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 405, 381–396 (2016). https://doi.org/10.1007/s11104-015-2495-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2495-4