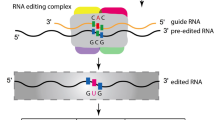
Abstract
RNA editing is post-transcriptional modification to RNA molecules. In plants, RNA editing primarily occurs to two energy-producing organelles: plastids and mitochondria. Organelle RNA editing is often viewed as a mechanism of correction to compensate for defects or mutations in haploid organelle genomes. A common type of organelle RNA editing is deamination from cytidine to uridine. Cytidine-to-uridine plastid RNA editing is carried out by the RNA editing complex which consists of at least four types of proteins: pentatricopeptide repeat proteins, RNA editing interacting proteins/multiple organellar RNA editing factors, organelle RNA recognition motif proteins, and organelle zinc-finger proteins. The four types of RNA editing factors work together to carry out RNA editing site recognition, zinc cofactor binding, and cytidine-to-uridine deamination. In addition, three other types of proteins have been found to be important for plastid RNA editing. These additional proteins may play a regulatory or stabilizing role in the RNA editing complex.
Similar content being viewed by others
Abbreviations
- APOBEC:
-
apolipoprotein B mRNA editing enzyme catalytic polypeptide-like
- CDA:
-
cytidine deaminase
- CLB:
-
chloroplast biogenesis
- CP31A:
-
31 kD chloroplast protein A
- CP31B:
-
31 kD chloroplast protein B
- CRR:
-
chlororespiratory reduction
- C-to-U:
-
cytidine-to-uridine
- DYW:
-
aspartate, tyrosine, and tryptophan domain
- E:
-
extension domain
- ECB:
-
early chloroplast biogenesis
- L:
-
long motif
- LPA:
-
low PSII accumulation
- MORF:
-
multiple organellar RNA editing factor
- OCP3:
-
overexpressor of cationic peroxidase
- ORRM:
-
organelle RNA recognition motif
- OTP:
-
organelle transcript processing
- OZ:
-
organelle zinc-finger
- P:
-
pentatricopeptide
- PPO/PPOX:
-
protoporphyrinogen oxidase
- PPR:
-
pentatricopeptide repeat
- QED:
-
quintuple editing factor
- RanBP:
-
Ran-binding-protein
- RARE:
-
required for accD RNA editing
- RIP:
-
RNA editing interacting protein
- RRM:
-
RNA recognition motif
- S:
-
short motif
- SMR:
-
PPR-small MutS-related
- THA:
-
thylakoid assembly
- T-to-C:
-
thymine-to-cytosine
- VAC:
-
vanilla cream
References
Barkan A., Small I.: Pentatricopeptide repeat proteins in plants. - Annu. Rev. Plant Biol. 65: 415–442, 2014.
Bentolila S., Heller W.P., Sun T. et al.: RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing.–P. Natl. Acad. Sci. USA 109: E1453–E1461, 2012.
Bentolila S., Oh J., Hanson M.R. et al.: Comprehensive highresolution analysis of the role of an Arabidopsis gene family in RNA editing.–PLoS Genet. 9: e1003584, 2013.
Betts L., Xiang S., Short S.A. et al.: Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex.–J. Mol. Biol. 235: 635–656, 1994.
Blokhina O., Fagerstedt K.V.: Reactive oxygen species and nitric oxide in plant mitochondria: origin and redundant regulatory systems.–Physiol. Plantarum 138: 447–462, 2010.
Blow M.J., Grocock R.J., van Dongen S. et al.: RNA editing of human microRNAs.–Genome Biol. 7: R27, 2006.
Boussardon C., Salone V., Avon A. et al.: Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids.–Plant Cell 24: 3684–3694, 2012.
Boussardon C., Avon A., Kindgren P. et al.: The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1.–New Phytol. 203: 1090–1095, 2014.
Brennicke A., Marchfelder A., Binder S.: RNA editing.–FEMS Microbiol. Rev. 23: 297–316, 1999.
Cai W., Ji D., Peng L. et al.: LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis.–Plant Physiol. 150: 1260–1271, 2009.
Cao Z.-L., Yu Q.-B., Sun Y. et al: A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development.–J. Integr. Plant Biol. 53: 258–269, 2011.
Chateigner-Boutin A.-L., Small I.: A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons.–Nucleic Acids Res. 35: e114, 2007.
Chateigner-Boutin A.L., Ramos-Vega M., Guevara-García A. et al.: CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts.–Plant J. 56: 590–602, 2008.
Chen M., Herde M., Witte C.P.: Of the nine cytidine deaminaselike genes in Arabidopsis, eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo.–Plant Physiol. 171: 799–809, 2016.
Chung S.J., Fromme J.C., Verdine G.L.: Structure of human cytidine deaminase bound to a potent inhibitor.–J. Med. Chem. 48: 658–660, 2005.
Coego A., Ramirez V., Gil M.J. et al.: An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens.–Plant Cell 17: 2123–2137, 2005.
Covello P.S., Gray M.W.: On the evolution of RNA editing. - Trends Genet. 9: 265–268, 1993.
Cui Y., Huang T., Zhang X.: RNA editing of microRNA prevents RNA-induced silencing complex recognition of target mRNA. - Open Biol. 5: 150126, 2015.
Delannoy E., Le Ret M., Faivre-Nitschke E. et al.: Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation.–Plant Cell 21: 2058–2071, 2009.
Faivre-Nitschke S.E., Grienenberger J.M., Gualberto J.M.: A prokaryotic-type cytidine deaminase from Arabidopsis thaliana gene expression and functional characterization.–Eur. J. Biochem. 263: 896–903, 1999.
Fey J., Weil J.H., Tomita K. et al.: Role of editing in plant mitochondrial transfer RNAs.–Gene 286: 21–24, 2002.
Foyer C.H., Harbinson J.: Oxygen metabolism and the regulation of photosynthetic electron transport.–In: Foyer C.H., Mullineaux P. (ed.): Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. Pp. 1–42. CRC Press, Boca Raton 1994.
Foyer C.H.: Oxygen metabolism and electron transport in photosynthesis.–In: Scandalios J. (ed.): Molecular Biology of Free Radical Scavenging Systems. Pp. 587–621. Cold Spring Harbor Laboratory Press, New York 1997.
García-Andrade J., Ramírez V., López A. et al.: Mediated plastid RNA editing in plant immunity.–PLOoS Pathog. 9: e1003713, 2013.
Gray M.W., Covello P.S.: RNA editing in plant mitochondria and chloroplasts.–FASEB J. 7: 64–71, 1993.
Hackett J.B., Shi X., Kobylarz A.T. et al.: An Organelle RNA Recognition Motif protein is required for photosystem II subunit psbF transcript editing.–Plant Physiol. 173: 2278–2293, 2017.
Hackett J.B., Lu Y.: Whole-transcriptome RNA-seq, gene set enrichment pathway analysis, and exon coverage analysis of two plastid RNA editing mutants.–Plant Signal. Behav. 12: e1312242, 2017.
Hammani K., Okuda K., Tanz S.K. et al: A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites.–Plant Cell 21: 3686–3699, 2009.
Hayes M.L., Giang K., Berhane B. et al.: Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. - J. Biol. Chem. 288: 36519–36529, 2013.
Hayes M.L., Dang K.N., Diaz M.F. et al: A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity.–J. Biol. Chem. 290: 10136–10142, 2015.
Hein A., Polsakiewicz M., Knoop V.: Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors.–BMC Evol. Biol. 16: 23, 2016.
Hirose T., Sugiura M.: Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system.–EMBO J. 20: 1144–1152, 2001.
Karcher D., Bock R.: Identification of the chloroplast adenosinetoinosine tRNA editing enzyme.–RNA 15: 1251–1257, 2009.
Kawahara Y., Zinshteyn B., Sethupathy P. et al.: Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. - Science 315: 1137–1140, 2007.
Ke J., Chen R.-Z., Ban T. et al.: Structural basis for RNA recognition by a dimeric PPR-protein complex.–Nat. Struct. Mol. Biol. 20: 1377–1382, 2013.
Kotera E., Tasaka M., Shikanai T.: A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts.–Nature 433: 326–330, 2005.
Kupsch C., Ruwe H., Gusewski S. et al.: Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps.–Plant Cell 24: 4266–4280, 2012.
Lynch M., Blanchard J.L.: Deleterious mutation accumulation in organelle genomes.–Genetica 102: 29, 1998.
Maris C., Dominguez C., Allain F.H.: The RNA recognition motif, a plastic RNA-binding platform to regulate posttranscriptional gene expression.–FEBS J. 272: 2118–2131, 2005.
Navaratnam N., Bhattacharya S., Fujino T. et al.: Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site.–Cell 81: 187–195, 1995.
Neiman M., Taylor D.R.: The causes of mutation accumulation in mitochondrial genomes.–P. Roy. Soc. B-Biol. Sci. 276: 1201–1209, 2009.
Nguyen C.D., Mansfield R.E., Leung W. et al.: Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA.–J. Mol. Biol. 407: 273–283, 2011.
Okuda K., Myouga F., Motohashi R. et al.: Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing.–P. Natl. Acad. Sci. USA 104: 8178–8183, 2007.
Okuda K., Chateigner-Boutin A.L., Nakamura T. et al.: Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts.–Plant Cell 21: 146–156, 2009.
Okuda K., Shoki H., Arai M. et al.: Quantitative analysis of motifs contributing to the interaction between PLS-subfamily members and their target RNA sequences in plastid RNA editing.–Plant J. 80: 870–882, 2014.
Robbins J.C., Heller W.P., Hanson M.R.: A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript.–RNA 15: 1142–1153, 2009.
Ruwe H., Castandet B., Schmitz-Linneweber C. et al.: Arabidopsis chloroplast quantitative editotype.–FEBS Lett. 587: 1429–1433, 2013.
Shi X., Bentolila S., Hanson M.R.: Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis.–Plant Signal. Behav. 11: e1167299, 2016.
Shi X., Hanson M.R., Bentolila S.: Functional diversity of Arabidopsis organelle-localized RNA-recognition motifcontaining proteins.–WIREs RNA 8: e1420, 2017.
Stern D.B., Goldschmidt-Clermont M., Hanson M.R.: Chloroplast RNA metabolism.–Annu. Rev. Plant Biol. 61: 125–155, 2010.
Sun T., Germain A., Giloteaux L. et al.: An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize.–P. Natl. Acad. Sci. USA 110: E1169–1178, 2013.
Sun T., Shi X., Friso G. et al: A zinc finger motif-containing protein is essential for chloroplast RNA editing.–PLoS Genet. 11: e1005028, 2015.
Sun T., Bentolila S., Hanson M.R.: The unexpected diversity of plant organelle RNA editosomes.–Trends Plant Sci. 21: 962–973, 2016.
Takenaka M., Zehrmann A., Verbitskiy D. et al.: Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants.–P. Natl. Acad. Sci. USA 109: 5104–5109, 2012.
Takenaka M., Zehrmann A., Verbitskiy D. et al: RNA editing in plants and its evolution.–Annu. Rev. Genet. 47: 335–352, 2013.
Tillich M., Hardel S.L., Kupsch C. et al.: Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs.–P. Natl. Acad. Sci. USA 106: 6002–6007, 2009.
Tseng C.C., Sung T.Y., Li Y.C. et al.: Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant.–Plant Mol. Biol. 73: 309–323, 2010.
Vincenzetti S., Cambi A., Neuhard J. et al.: Cloning, expression, and purification of cytidine deaminase from Arabidopsis thaliana.–Protein Expr. Purif. 15: 8–15, 1999.
Wagoner J.A., Sun T., Lin L. et al.: Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeatcontaining proteins are required for site-specific chloroplast RNA editing.–J. Biol. Chem. 290: 2957–2968, 2015.
Xu J.-H., Messing J.: Maize haplotype with a helitron-amplified cytidine deaminase gene copy.–BMC Genetics 7: 1–13, 2006.
Yagi Y., Tachikawa M., Noguchi H. et al.: Pentatricopeptide repeat proteins involved in plant organellar RNA editing. - RNA Biol. 10: 1419–1425, 2013.
Yang W., Chendrimada T.P., Wang Q. et al.: Modulation of microRNA processing and expression through RNA editing by ADAR deaminases.–Nat. Struct. Mol. Biol. 13: 13–21, 2006.
Yap A., Kindgren P., Colas des Francs-Small C. et al.: AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice.–Plant J. 81: 661–669, 2015.
Yin P., Li Q., Yan C. et al.: Structural basis for the modular recognition of single-stranded RNA by PPR proteins.–Nature 504: 168–171, 2013.
Yu Q.-B., Jiang Y., Chong K. et al.: AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana.–Plant J. 59: 1011–1023, 2009.
Zehrmann A., Härtel B., Glass F. et al.: Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana.–J. Biol. Chem. 290: 6445–6456, 2015.
Zhang F., Tang W., Hedtke B. et al.: Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing.–P. Natl. Acad. Sci. USA 111: 2023–2028, 2014.
Zhou W., Cheng Y., Yap A. et al.: The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth.–Plant J. 58: 82–96, 2009.
Zhou W., Karcher D., Bock R.: Identification of enzymes for adenosine-to-inosine editing and discovery of cytidine-touridine editing in nucleus-encoded transfer RNAs of Arabidopsis.–Plant Physiol. 166: 1985–1997, 2014.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgments: This work was supported by the U.S. National Science Foundation (grant no. MCB-1244008).
Rights and permissions
About this article
Cite this article
Lu, Y. RNA editing of plastid-encoded genes. Photosynthetica 56, 48–61 (2018). https://doi.org/10.1007/s11099-017-0761-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11099-017-0761-9