Abstract
The human brain is organized into functional networks, whose spatial layout can be described with functional magnetic resonance imaging (fMRI). Interactions among these networks are highly dynamic and nonlinear, and evidence suggests that distinct functional network configurations interact on different levels of complexity. To gain new insights into topological properties of constellations interacting on different levels of complexity, we analyze a resting state fMRI dataset from the human connectome project. We first measure the complexity of correlational time series among resting state networks, obtained from sliding window analysis, by calculating their sample entropy. We then use graph analysis to create two functional representations of the network: A ‘high complexity network’ (HCN), whose inter-node interactions display irregular fast changes, and a ‘low complexity network’ (LCN), whose interactions are more self-similar and change more slowly in time. Graph analysis shows that the HCNs structure is significantly more globally efficient, compared to the LCNs, indicative of an architecture that allows for more integrative information processing. The LCNs layout displays significantly higher modularity than the HCNs, indicative of an architecture lending itself to segregated information processing. In the HCN, subcortical thalamic and basal ganglia networks display global hub properties, whereas cortical networks act as connector hubs in the LCN. These results can be replicated in a split sample dataset. Our findings show that investigating nonlinear properties of resting state dynamics offers new insights regarding the relative importance of specific brain regions to the two fundamental requirements for healthy brain functioning, that is, integration and segregation.
Similar content being viewed by others
1 Introduction
Fluctuations of the blood oxygen level-dependent (BOLD) signal from the human brain display spatiotemporally organized patterns of functional connectivity (FC) during rest, forming so-called resting state networks (RSNs) [1, 2]. Recently, there has been increased interest in the temporal dynamics of FC [3, 4], which have been quantified with complexity measures (inter alia), like approximate entropy (AE) [5], Hurst exponent [6], and sample entropy (SampEn) [7]. In studies relevant to the present investigation, the nonlinear dynamics of correlational time series (CTS), obtained with a sliding window (SW) [8], were assessed by computing their SampEn [9,10,11]. These studies were either focused on differences in SampEn between schizophrenic patients and healthy controls [10, 11], or the spatial overlap between the SampEn gradient across the brain and established RSNs [9]. However, this methodology has yet to be employed to investigate topological features of different network constellations, dissociated based on the degree of nonlinearity governing the interactions among their constituent parts.
We therefore combined this approach with graph-theoretic measures (GMs) used extensively to describe topological features of brain networks [12, 13]. In most graph-based approaches, the edges/connections between two given nodes (corresponding to brain regions) to be included in a graph are selected according to correlation-strength, and the edges falling below some cutoff are being discarded [14]. Here, we created two subgraphs of the fully connected graph, with nodes corresponding to RSNs, and the edges to the SampEn of the CTS between them. Our choice to use SampEn as a complexity index was motivated by the fact that it has been used extensively as an analysis tool for physiological time series (TS) [15,16,17,18]. More importantly for the purposes of our study, its parameter choices (see Sect. 2.5) have been validated for (raw) BOLD time series [7] and CTS derived from SW analysis [9,10,11].
We kept edges with the highest and lowest SampEn, respectively, to obtain a high complexity graph (HCG) and a low complexity graph (LCG). Compatible with the formal definition of SampEn, our subgraphs can be interpreted as representations of distinct functional configurations of the same network forming at different temporal scales: Higher SampEn of the CTS among two given nodes indicates faster dynamics, reflected by the irregularity of the CTS. On the other hand, lower SampEn (more self-similarity of the CTS) corresponds to slower dynamics. This procedure was motivated by the fact that the importance of a node in a brain network (its ‘hubness’) seems to be frequency-dependent [19, 20], and depends on the temporal resolution of the methodology employed [21]. In line with these and further results indicating the temporal specificity of intra- and inter-RSN interactions [22,23,24], we expected significant differences in topology between HCGs and LCGs, at the node as well as the global level.
To test our hypothesis, we used resting state data of healthy subjects from the human connectome project (HCP) [25].
Short summary:
-
Human connectome project resting state time series
-
Sliding window analysis of the BOLD time series
-
Sample entropy calculation of the sliding window time series
-
Derivation of high complexity and low complexity graphs from the sample entropy matrices
-
Topological analysis with global and local graph measures
2 Methods
2.1 Participants
The whole sample was made up of 812 (N = 812) (410 women; n = 410) healthy subjects from the HCP between the ages of 22 to 37 years. The sample was then split into two sets of 406 participants each to obtain a replication dataset.
2.2 Resting state fMRI data
The subjects imaging data consisted of a subset of the high-level resting state connectivity analyses outputs, based on the 2017 HCP data release. BOLD TS contained 2400 time-points, corresponding to two concatenated 15 min sessions, to control for any influences of the phase-encoding direction, which was left to right, and right to left, respectively, for the two sessions. The data were collected with the following parameters: gradient-echo EPI sequence, TR = 720 ms, TE = 33.1 ms, flip angle = 52°, field of view = 208 × 180 (RO x PE), matrix 104 × 90 (RO x PE), slice thickness = 2 mm, 72 slices, 2.0 mm isotropic voxels, multiband factor = 8, echo spacing 0.58 ms, and bandwidth = 2290 Hz/Px. Then data were (pre)processed according to the minimal preprocessing pipeline of the HCP [26, 27] as well as subjected to artifact removal [28, 29]. Subsequently, a group-based principal component analysis (PCA) was carried out [30]. The group PCA output was used as input for group ICAs to obtain parcellations at different dimensionalities based on data from the whole sample [27]. This was done with the help of FSL’s MELODIC toolbox [31, 32]. Finally, individual TS for the ICA components were reconstructed with a dual regression technique [33], resulting in one representational BOLD TS per ICA component for every subject.
2.3 RSN selection
The RSN selection from the 50 components was carried out in an automatized way by using the component labeler of the GIFT toolbox (https://trendscenter.org/software/gift/), which uses the templates from the Functional Imaging in Neuropsychiatric Disorders Lab at Stanford University, USA (https://findlab.stanford.edu/functional_ROIs.html) [34]. Each components’ spatial map was correlated with these templates and subsequently assigned to the best fitting template (i.e., the corresponding RSN), based on the maximum correlation value (only fits above r = 0.15 were included). This resulted in 23 (N = 23) cortical/subcortical components, corresponding to the following networks (NW): Auditory NW (AUD; n = 1), basal ganglia NW (BG; n = 1), sensorimotor NW (SM; n = 2), visual NW (VIS; n = 5), visuospatial NW (VSP; n = 2), DMN (n = 2), executive control NW (EXC; n = 3), language NW (LNG; n = 2), precuneus NW (PREC; n = 1), and salience NW (SAL; n = 3). VIS was further divided into one pVIS (n = 1) and four higher visual NWs (hVIS; n = 4). The DMN was split up into one dorsal (dDMN; n = 1) and one ventral (vDMN; n = 1) subcomponent. The EXC was partitioned into one right-lateralized (rEXC; n = 1) and two left-lateralized NWs (lEXC; n = 2). Finally, SAL was divided into two anterior NWs (aSAL; n = 2) as well as one posterior NW (pSAL; n = 1). Components with extensive cerebellar activations were excluded from the analysis.
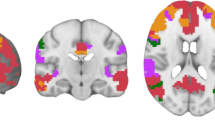
The component labeled vDMN by the GIFT toolbox (r = 0.39) also displayed considerable activations in the precuneus and was therefore denoted vDMN/PC, upon visual inspection. The component labeled PRE by the GIFT toolbox (r = 0.33) displayed considerable activations in the cuneus and was therefore denoted PREC/CUN. One component was identified as a thalamic NW (TH; n = 1) upon visual inspection and included in the analysis as such. The abbreviations for the components associated with their corresponding RSNs will be used as proxies for the components from now on. For example, vDMN/PC stands for the IC associated with the vDMN/PC. For a depiction of the RSNs spatial maps, see Fig. 1.
Components chosen from the results of the independent component analysis with dimensionality 50 (ICA-50). ICA maps were dual regressed into subjects 3d data and then averaged across subjects; the images show the activations (red) at the most relevant axial, coronal, and sagittal slices of Montreal Neurological Institute (MNI) 152 space. Below the slices are the corresponding resting state networks and components: Auditory network (AUD; n = 1), basal ganglia network (BG; n = 1), thalamic network (TH; n = 1), sensorimotor networks (SM; n = 2), visuospatial networks (VSP; n = 2), language networks (LNG; n = 2), precuneus/cuneus network (PREC/CUN, n = 1), primary visual network (pVIS; n = 1), higher visual networks (hVIS; n = 4), dorsal default mode network (dDMN; n = 1), ventral default mode/precuneus network (vDMN/PC; n = 1), right executive control network (rEXC; n = 1), left executive control network (lEXC; n = 2), anterior salience network (aSAL; n = 2), and posterior salience network (pSAL; n = 1). (Color figure online)
2.4 Sliding window
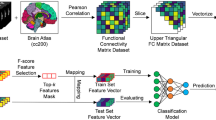
We used a 60-s tapered window (rectangle convolved with a Gaussian: standard deviation [SD] = 1TR), after filtering the BOLD TS from 0.017 to 0.1 Hz to remove frequencies lower than 1/w, where w is the window size. This was done to limit the influence of temporal spurious fluctuations in FC [35]. The window was slid forward in steps of 1TR, resulting in a series of 2317 correlation matrices of size 23 × 23, i.e., in 253 (23 × 22/2) CTS. See Fig. 2 for a graphical description of the workflow described in Sects. 2.4–2.6.
Graphical description of the workflow for a single subject. a: A 60-s tapered window was slid over the filtered BOLD time series (TS) to obtain the time-resolved functional connectivity (FC) estimates between every pair of regions. b: The sample entropy was calculated for every FC TS. c, d: From the resulting sample entropy matrix two binary adjacency matrices were created, by keeping the edges (connections) with the highest and lowest sample entropy values, respectively. e: Two undirected graphs (high complexity, and low complexity) were created from the adjacency matrices and subjected to further analysis. (Color figure online)
2.5 Sample entropy
To compute the SampEn of a given CTS x = [ \({x}_{1},{x}_{2,\cdots },{x}_{n}\)] with length N, one creates an embedding vector with m running data points from x: \({v}_{i}=\left[{x}_{i},{x}_{i+1},\dots ,{x}_{i+m-1}\right]\), with m being the embedding dimension. Then, for each i (1 ≤ i ≤ N – m) define
where r = εσx is a tolerance value, ε a scaling parameter, and σx the SD of x. Θ(·) is the Heaviside function.
and \({\Vert \cdot \Vert }_{1}\) is the Chebyshev distance, i.e.,
Subsequently, for each i (1 ≤ i ≤ N – m) define.
Averaging over all m embedding vectors, we have that.
and
Then, the SampEn is defined as.
resulting in a nonnegative number, with higher values indicative of more complexity, and lower values indicative of more regularity of the underlying CTS. We did this for every CTS of every subject, resulting in a 406 × 253 SampEn matrix. We choose the standard parameter values that are used in the literature, i.e., m = 2 and ε = 0.20 [9, 10, 36], to ensure comparability of our results with these past investigations.
To validate our choice of complexity measure (SampEn), we also calculated an alternative, amplitude based, complexity index, namely AE (for a detailed description/derivation of AE, see [36, 37]). Subsequently, the 253 SampEn indices were correlated with their corresponding AE counterparts for every subject, with Pearson’s r. The correlations were generally high (range: 0.77–0.93, mean correlation: r = 0.87 [first fisher-z and then back transformed]), and statistically significant for every subject (Bonferroni corrected), as well as statistically different from zero (one-sample T test).
Since basic BOLD signal properties can influence complexity measures [38], we calculated the temporal signal-to-noise ratio (tSNR) (mean relative to SD) from the (pre-processed) unfiltered BOLD TS, as well as for the filtered version (bandpass from 0.017 to 0.1 Hz) for every RSN (node), for every subject. These values were then correlated with the 253 SampEn values of the CTS with Pearson’s r. There was only one correlation among SampEn and tSNR that reached significance (Bonferroni corrected), namely between hVIS4 and the SampEn for the CTS hVIS4 aSAL1, with r(404) = − 0.16, p < 0.001.
2.6 Graph measures
To create the two graphs of the network (high complexity vs. low complexity), we binarized the SampEn matrices after selecting the highest (HCG) and lowest (LCG) SampEn connections, respectively. To avoid that the values of our graph measures (GMs) would be biased by a specific threshold, we computed all GMs for a range of density values. The reported results were averaged across thresholds, following the approach for correlation-based graph analyses suggested by [39]. For a graphical description of our workflow, see Fig. 2.
2.6.1 Global efficiency
Global efficiency (GE) is an index measuring the efficiency of information exchange in a network, i.e., its functional integration [40], and is defined as the inverse average shortest path length:
where \({d}_{ij}\) represents the shortest path from node i to node j [41]. We computed the average GE of every subject, for the HCG and LCG, respectively. We then compared their means with a T test for dependent samples.
2.6.2 Betweenness centrality
To assess the importance of a given node, we computed its betweenness centrality (BC) [14], defined by the following expression:
where \({\sigma }_{ij}\) is the total number of shortest paths from node i to node j, \({\sigma }_{ij}\left(v\right)\) is the number of those paths passing through v, and n is the number of nodes in the graph. We did this for all 23 nodes of the HCG and LCG, respectively, for every subject. As null models we computed 1000 random graphs with the same node degree distribution [42], for the HCG and LCG of every subject. Following a procedure described by [12], we then averaged the BC values of the 23 nodes across the 1000 graphs across all subjects and subtracted this cutoff value from the average BC of every node from the non-random graphs. A node was determined to be a hub if this value was at least one SD above the average difference across all nodes. This way we obtained the hub structure for the HCG and LCG, respectively.
2.6.3 Modularity
As a measure of functional segregation, we computed the Newman–Girwan modularity (MOD) of the graphs, which, given a partition of the network nodes, results in an index Q (if Q = 0 the community structure of the graph is not stronger than one would expect by chance; Q > 0 if stronger, and Q < 0 if weaker) [43]. To obtain the optimal partition and deal with the known issue of degeneracy [44], we used the Louvain algorithm [45], as part of a procedure described in [46]: After running the algorithm 100 times, we computed the (23 × 23) association matrix, where entry \({A}_{ij}\) represents the number of times node i and node j are assigned to the same community across the 100 iterations. From this matrix, we subtracted a null model association matrix obtained from random permutations of the original partitions and kept the values above zero. Finally, we obtained the final partition by running the algorithm on the resulting association matrix. We computed Q for the HCG and LCG for every subject and subsequently compared their means with a T test for dependent samples. To gain more insight into the properties of the hubs, we obtained with the procedure described in 2.6.2 we calculated the within-module z-score (WMZ). The WMZ value of node i is defined as:
where \({m}_{i}\) is the module of node i, \({k}_{i}\left({m}_{i}\right)\) the within-module degree of i (the number of edges between i and all other nodes in \({m}_{i}\)), and \(\overline{k }\left({m}_{i}\right)\) and \({\sigma }^{k\left({m}_{i}\right)}\) are the mean and SD of the within-module \({m}_{i}\) degree distribution [47]. We also computed the participation coefficient (PC), which, for node i, is equivalent to:
where M is the set of modules (given by the partition of the nodes), and \({k}_{i}\left(m\right)\) is the number of edges between i and all nodes in module m [47]. Nodes with high WMZ score and low PC contribute mainly to the modular segregation of a network (provincial hub [PH]), whereas a node with high PC contributes to (inter-modular) integration (connector hub [CH]) [41]. Subsequently, we performed multiple comparisons of these measures between our hubs (HCG and LCG).
2.7 Software used for analyses and visualization
All calculations were performed with MATLAB (9.7.0.1296695 [R2019b] Natick, Massachusetts: The MathWorks Inc.). Figures 1 and 2 were in part created with MATPLOTLIB [48]. Figures 1 and 3 were in part created with the help of the visualization tool SCHEMABALL (https://habs.mgh.harvard.edu/researchers/data-tools/schemaball).
Upper panel: Boxplots of the global efficiency values (left side), and modularity Q values (right side), for the high complexity network (red boxes) and the low complexity network (blue boxes), respectively. The black line within the boxes represents the median, the area of a box covers the inter-quartile range (IQR) for the data values. Whiskers indicate the range of the data, and individual points show values higher than the third quartile + 1.5*IQR, and lower than the first quartile − 1.5*IQR, respectively. Lower panel: Bar plots of the betweenness centrality (BC) values for the networks in the high complexity network (left side; red), and the low complexity network (right side; blue): Nodes with significant BC values are marked by an asterisk. Significance for a node was reached when its average BC across all subjects was at least one standard deviation above the average difference between the BC values across all nodes and subjects, and the average BC value across all nodes and subjects derived from a series of random graphs, preserving the node degree distribution. AUD = auditory network; BG = basal ganglia network; TH = thalamic network; SM1, SM2 = sensorimotor networks; VSP1, VSP2 = visuospatial networks; LNG1, LNG2 = language networks; PREC/CUN = precuneus/cuneus network; pVIS = primary visual network; hVIS1, hVIS2, hVIS3, hVIS4 = higher visual networks; dDMN = dorsal default mode network; vDMN = ventral default mode network; rEXC1 = right executive control network; lEXC1, lEXC2 = left executive control networks; aSAL1, aSAL2 = anterior salience networks; pSAL = posterior salience network. (Color figure online)
3 Results
3.1 Graph measures
3.1.1 Global efficiency
Average GE of the HCGs (M = 0.59, SEM = 0.005) was significantly higher, compared to the average GE of the LCGs (M = 0.58, SEM = 0.009), with t(405) = 9.15, p < 0.001, and Cohen’s d = 0.45 (medium size effect), see Fig. 3.
3.1.2 Betweenness centrality/hubs
In the HCG BG (M = 0.09; SD = 0.07) and TH (M = 0.10; SD = 0.07) displayed significant BC. In the LCG the following nodes displayed significant BC: SM2 (M = 0.06; SD = 0.04), hVIS1 (M = 0.05; SD = 0.04), LNG1 (M = 0.05; SD = 0.04), and VSP2 (M = 0.06; SD = 0.04). To compare the hubs across the HCGs and LCGs in terms of their BC, we used a nonparametric Friedman test for repeated measures, which resulted in a significant column effect (Chi-square value = 139.24, p < 0.001, df = 5). Multiple comparisons (Bonferroni corrected) revealed that BG and TH had significantly higher mean ranks (BG = 4.07; TH = 4.20), compared to the hubs of the LCGs, which did not differ significantly in their mean rank, with SM2 = 3.26, hVIS1 = 3.13, LNG1 = 3.13, and VSP2 = 3.24. See Figs. 3 and 4 for a graphical description of the results reported in this section.
Graphical description of the topology of the high complexity network (left; red) and the low complexity network (right; blue) at a density of 18 percent, i.e., with edges representing the top (high complexity network) and bottom (low complexity network) 18 percent functional connectivity time series in terms of sample entropy. Hubs are surrounded by a black box. Note: The graphs were created by averaging the node degrees across all subjects and then rounding to the nearest integer. Subsequently a representational graph was created with the Havel–Hakimi algorithm [85, 86]. This procedure was not part of any calculations to obtain the results of this study. AUD = auditory network; BG = basal ganglia network; TH = thalamic network; SM1, SM2 = sensorimotor networks; VSP1, VSP2 = visuospatial networks; LNG1, LNG2 = language networks; PREC/CUN = precuneus/cuneus network; pVIS = primary visual network; hVIS1, hVIS2, hVIS3, hVIS4 = higher visual networks; dDMN = dorsal default mode network; vDMN = ventral default mode network; rEXC1 = right executive control network; lEXC1, lEXC2 = left executive control networks; aSAL1, aSAL2 = anterior salience networks; pSAL = posterior salience network. (Color figure online)
3.1.3 Modularity/hubs
Average Q of the LCGs (M = 0.49, SEM = 0.002) was significantly higher, compared to the average Q of the HCGs (M = 0.48, SEM = 0.002), with t(405) = 3.34, p < 0.001, and Cohen’s d = 0.17 (small size effect), see Fig. 2. We also statistically compared the average number of clusters, which was significantly higher for the LCGs (M = 3.49, SEM = 0.03), compared to the HCGs (M = 3.34, SEM = 0.02), with t(405) = 4.07, p < 0.001, and Cohen’s d = 0.20 (small size effect).
In terms of PC all identified hubs can be considered CHs, if one uses the cutoff of 0.3 as suggested by [47]: BG (M = 0.55; SD = 0.11), TH (M = 0.55; SD = 0.10), SM2 (M = 0.49; SD = 0.15), hVIS1 (M = 0.48; SD = 0.15), LNG1 (M = 0.50; SD = 0.14), and VSP2 (M = 0.51; SD = 0.12). As one would expect, all hubs had positive WMZ scores: BG (M = 0.58; SD = 0.72), TH (M = 0.60; SD = 0.70), SM2 (M = 0.25; SD = 0.67), hVIS1 (M = 0.25; SD = 0.64), LNG1 (M = 0.19; SD = 0.61), and VSP2 (M = 0.20; SD = 0.64). As a cutoff, we defined that for a hub to be considered a PH its WMZ score had to be at least a SD above the mean WMZ score across all hubs, which was the case for BG and TH only.
To compare the hubs in terms of their PC we used a nonparametric Friedman test for repeated measures, which resulted in a significant column effect (Chi-square value = 132.27, p < 0.001, df = 5). Multiple comparisons (Bonferroni corrected) revealed that BG and TH had significantly higher mean ranks (BG = 4.11; TH = 4.09), compared to the other hubs, which did not differ significantly in their mean rank, with SM2 = 3.13, hVIS1 = 3.04, LNG1 = 3.27, and VSP2 = 3.36. To compare the hubs in terms of their WMZ we used a nonparametric Friedman test for repeated measures, which resulted in a significant column effect (Chi-square value = 131.76, p < 0.001, df = 5). Multiple comparisons (Bonferroni corrected) revealed that BG and TH had significantly higher mean ranks (BG = 4.07; TH = 4.13), compared to the other hubs, which did not differ significantly in their mean rank, with SM2 = 3.32, hVIS1 = 3.31, LNG1 = 3.07, and VSP2 = 3.11.
3.2 Replication data set and additional analyses
In the (split sample) replication data set, all results from the original set could be replicated. We also wanted to assess if our main results were independent of our method to derive RSNs (dual regression, automatized assignment of ICA-derived components to specific functional NWs), and our decision to binarize the SampEn matrices in deriving HCGs and LCGs. We therefore repeated our analysis on a subset (n = 100) of subjects after choosing a different (a priori) parcellation, based on [49], and subsequently computed weighted versions of our main GMs (GE, MOD, and BC). The resulting effect sizes were equivalent in direction, and either larger (GE; Cohen’s d > 0.80, large size effect), or analogous (MOD; Cohen’s d = 0.18, small size effect), compared to the ones from the main analyses. In terms of BC, thalamic and basal ganglia regions retained their hub status in the HCGs, as did higher visual, sensorimotor, and attentional (akin to VSP2) regions in the LCGs. We therefore concluded that our original results were unbiased by subject selection, parcellation, and binarization of the SampEn matrices.
4 Discussion
4.1 Global efficiency and modularity
The main finding of our investigation is that when RSNs interact with high levels of complexity and irregularity they display a topology that permits more efficient integration of information, compared to when their interplay is governed by lower levels of complexity and changes at a slower pace. On the other hand, the low complexity network (LCN) shows a higher degree of functional segregation, compared to the high complexity network (HCN). Importantly, this pattern of results is replicable across datasets, parcellations, and different ways to derive the HCGs and LCGs from the SampEn matrices. These outcomes are in line with past investigations reporting topological differences of functional RSN configurations at distinct frequencies [19, 20, 50]. Our outcomes are also compatible with evidence from fMRI that the brain during rest switches between periods where its network structure exhibits different degrees of MOD [51], and with evidence based on magnetoencephalography that the same holds true for GE [12]. Since our results were obtained with a (to our knowledge) new combination of methodological approaches (namely complexity analysis of CTS and graph-theoretic measures), they underscore the validity of the studies mentioned above, in the sense of multiple operationalization. Additionally, they could open the door to gaining new insights into the mechanisms underlying mental illness (see Sect. 4.3 for a discussion).
4.2 Hub structure
BG and TH emerged consistently as hubs of the HCN across datasets, and both qualified as a CH as well as a PH, according to our criteria. It has been suggested that TH acts as global/kinless hub capable of interacting with multiple cortical networks [47, 52, 53], allowing for efficient and integrative information processing [54, 55]. Of note, age-related decreases in GE are related to decreased local efficiency of BG and TH structures [56], and, in accordance with its global hub properties in our study, lesions to TH also result in decreased MOD on a network level [39]. The structural basis for the CH properties of TH and BG is well established, of course as constituent elements of the cortico-basal ganglia-thalamo-cortical loop [57, 58], but also through extensive reciprocal interconnections of TH (mainly with prefrontal cortex) through which it can exert influence on cortico-cortical activity [59, 60].
When compared to the HCN, the overall structure of the LCN seemed to be less determined by properties of a few hubs, since PPC and BC values of the LCN hubs were significantly lower than for BG and TH in the HCN. The average number of clusters was significantly higher in the LCN, as was the average MOD, indicative of a topology that favors functional segregation, juxtaposed with the HCN. The spatial layout of most LCN hubs encompassed regions that have consistently been associated with networks exhibiting functional hub properties [12, 61,62,63]. These were precentral and postcentral gyri (SM2), cuneus (hVIS1), and inferior parietal lobule (VSP2). Interestingly, a similar hub structure emerges when the graph-analyses are based on bandpass-filtered TCs (between 0.01 and 0.075 Hz) [19], although global measures of network topology were not evaluated in this study. This temporal dimension of the (functional) network architecture of the brain is also reflected in our results.
4.3 Clinical applications
Our results and methodology can potentially offer new insights concerning neuronal correlates of neuropsychiatric illnesses, such as major depressive disorder (MDD) and schizophrenia (SCZ). With regard to MDD, numerous studies have found diverging FC between patients and healthy controls [64,65,66], but often the temporal specificity of these patterns is not taken into account. However, it has been shown that differences in hub structure between MDD and healthy populations are a function of the dynamics with which these brain regions interact [19]. In light of our results, it would be interesting to investigate changes that are specific to the HCN and LCN, respectively, at the global (GE and MOD), as well as the node level (e.g., the hub structure). Since MDD populations are not homogenous in terms of their symptoms and impairments [67], one can speculate that a HCN/LCN specific analysis could shed light on the physiological underpinnings of these different manifestations of the disease; however, testing these hypotheses has to be the focus of future investigations.
The same holds true for the study of SCZ patients, where affective and psychotic symptoms can affect subpopulations to a different degree [67, 68]. Additionally, given that BG dysfunction in SCZ has been established by a plethora of studies and meta-analyses [69,70,71], the global hub properties of BG found in the present investigation would predict decreased GE, as well as MOD, in SCZ patients, compared to controls. There is indeed evidence that this seems to be the case [72,73,74,75], and interestingly, TH dysfunction has also been found in SCZ [76, 77], which is also in line with our results. Of note, after calculating the SampEn of CTS obtained from a SW (analogous to our study), [11] reported increased complexity within the visual system of SCZ patients, as well as within and between subcortical and cortical structures [10]. In light of our results, and the findings mentioned above, one can speculate that this might be reflective of a compensatory mechanism [78], to account for decreased effectiveness of BG and TH in ensuring proper information processing. Apart from MDD and SCZ, we think that our approach in general can offer new insights to the workings of the healthy, aging, and diseased brain, and opens the door for future investigations of nonlinear brain network interactions across different populations.
4.4 Limitations
There are some caveats inherent to our approach. First, the use of a SW to assess FC dynamics requires determining the values of two parameters (window size and SD) a priori; however, a substantial body of literature exists that suggests that our choices are close to optimal for BOLD time series [35, 79,80,81]. It should be noted that a number of other ways for estimating dynamic FC exist, such as spatial distance, innovation-driven co-activation patterns, and jackknife correlation [82,83,84], that offer a frame by frame temporal resolution, whereas (by its nature), SW analysis rather captures slower FC dynamics. Applying our approach to these alternative methods is an exciting prospect for future investigations. Secondly, in terms of our graph analyses, we concede that the choice of the cutoff values (determining the density of the graphs) is somewhat arbitrary, but by averaging the GMs across a range of cutoff values we are confident that our results are not biased by a specific threshold. Finally, another issue pertains to the influence of noise. By definition, SampEn is sensitive to noise in two ways: Unstructured noise would result in higher SampEn values, whereas structured noise would deflate SampEn values. Nevertheless, the fact that SampEn values were generally not significantly correlated with the tSNR of their BOLD TS indicates that they were not simply a function of the noise content of the underlying TS.
5 Summary
We have shown that subcortical networks are the central nodes in a large-scale brain network whose inter-node interactions are marked by fast dynamics and high levels of complexity. The topology of this HCN favors the integration of information, juxtaposed to a LCN governed by slower dynamics. In contrast, the LCNs structure displays higher levels of MOD than the HCN. These results are important, since they show that investigating nonlinear properties of resting state dynamics can reveal the relative importance of specific brain regions to different fundamental requirements for healthy brain functioning.
Data availability
The HCP data are freely available at http://www.humanconnectomeproject.org/data/.
References
Damoiseaux, J.S., et al.: Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U.S.A. 103(37), 13848–13853 (2006)
De Luca, M., et al.: fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage 29(4), 1359–1367 (2006)
Lurie, D.J., et al.: Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Network Neuroscience 4(1), 30–69 (2020)
Calhoun, V.D., et al.: The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron 84(2), 262–274 (2014)
Liu, C.Y., et al.: Complexity and synchronicity of resting state blood oxygenation level-dependent (BOLD) functional MRI in normal aging and cognitive decline. J. Magn. Reson. Imag. 38(1), 36–45 (2013)
Dong, J., et al.: Hurst exponent analysis of resting-state fMRI signal complexity across the adult lifespan. Front. Neurosci. 12, 34 (2018)
Yang, A.C., et al.: A strategy to reduce bias of entropy estimates in resting-state fMRI signals. Front. Neurosci. 12, 398 (2018)
Hutchison, R.M., et al.: Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp 34(9), 2154–2177 (2013)
Jia, Y., Gu, H.: Sample entropy combined with the K-means clustering algorithm reveals six functional networks of the brain. Entropy 21(12), 1156 (2019)
Jia, Y., Gu, H., Luo, Q.: Sample entropy reveals an age-related reduction in the complexity of dynamic brain. Sci. Rep. 7(1), 7990 (2017)
Jia, Y., Gu, H.: Identifying nonlinear dynamics of brain functional networks of patients with schizophrenia by sample entropy. Nonlinear Dyn. 96(4), 2327–2340 (2019)
de Pasquale, F., et al.: A dynamic core network and global efficiency in the resting human brain. Cereb Cortex 26(10), 4015–4033 (2016)
van den Heuvel, M.P., Sporns, O.: An anatomical substrate for integration among functional networks in human cortex. J. Neurosci. 33(36), 14489–14500 (2013)
Fornito, A., Zalesky, A., Breakspear, M.: Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage 80, 426–444 (2013)
Alcaraz, R., et al.: Study of Sample Entropy ideal computational parameters in the estimation of atrial fibrillation organization from the ECG. vol. 37, 1027–1030 (2010).
Richman, J.S., Moorman, J.R.: Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circul. Physiol. 278(6), H2039–H2049 (2000)
Keller, K., et al.: Permutation entropy: new ideas and challenges. Entropy 19(3), 134 (2017)
Cuesta-Frau, D.: Permutation entropy: Influence of amplitude information on time series classification performance. Math Biosci Eng 16(6), 6842–6857 (2019)
Ries, A., et al.: Frequency-dependent spatial distribution of functional hubs in the human brain and alterations in major depressive disorder. Front Hum Neurosci 13, 146 (2019)
Thompson, W.H., Fransson, P.: The frequency dimension of fMRI dynamic connectivity: network connectivity, functional hubs and integration in the resting brain. Neuroimage 121, 227–242 (2015)
Fransson, P., Thompson, W.H.: Temporal flow of hubs and connectivity in the human brain. Neuroimage 223, 117348 (2020)
Sasai, S., et al.: Frequency-specific network topologies in the resting human brain. Front. Hum. Neurosci. 8, 1022 (2014)
Salvador, R., et al.: A simple view of the brain through a frequency-specific functional connectivity measure. Neuroimage 39(1), 279–289 (2008)
Handwerker, D.A., et al.: Periodic changes in fMRI connectivity. Neuroimage 63(3), 1712–1719 (2012)
Van Essen, D.C., et al.: The WU-Minn human connectome project: an overview. Neuroimage 80, 62–79 (2013)
Smith, S.M., et al.: Resting-state fMRI in the human connectome project. Neuroimage 80, 144–168 (2013)
Glasser, M.F., et al.: The minimal preprocessing pipelines for the human connectome project. Neuroimage 80, 105–124 (2013)
Salimi-Khorshidi, G., et al.: Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 90, 449–468 (2014)
Griffanti, L., et al.: ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage 95, 232–247 (2014)
Smith, S.M., et al.: Group-PCA for very large fMRI datasets. Neuroimage 101, 738–749 (2014)
Hyvarinen, A.: Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 10(3), 626–634 (1999)
Beckmann, C.F., Smith, S.M.: Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imag. 23(2), 137–152 (2004)
Filippini, N., et al.: Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. U S A 106(17), 7209–7214 (2009)
Shirer, W.R., et al.: Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22(1), 158–165 (2012)
Leonardi, N., Van De Ville, D.: On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage 104, 430–436 (2015)
Pincus, S.M., Goldberger, A.L.: Physiological time-series analysis: what does regularity quantify? Am. J. Physiol. 266(4 Pt 2), H1643–H1656 (1994)
Pincus, S.M., Gladstone, I.M., Ehrenkranz, R.A.: A regularity statistic for medical data analysis. J. Clin. Monit. 7(4), 335–345 (1991)
Keilholz, S., et al.: Relationship between basic properties of BOLD fluctuations and calculated metrics of complexity in the human connectome project. Front. Neurosci. 14, 939 (2020)
Hwang, K., et al.: The human thalamus is an integrative hub for functional brain networks. J Neurosci 37(23), 5594–5607 (2017)
Latora, V., Marchiori, M.: Efficient behavior of small-world networks. Phys. Rev. Lett. 87(19), 198701 (2001)
Rubinov, M., Sporns, O.: Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52(3), 1059–1069 (2010)
Maslov, S., Sneppen, K.: Specificity and stability in topology of protein networks. Science 296(5569), 910–913 (2002)
Newman, M.E.J.: Detecting community structure in networks. Eur. Phys. J. B 38(2), 321–330 (2004)
Good, B.H., de Montjoye, Y.A., Clauset, A.: Performance of modularity maximization in practical contexts. Phys. Rev. E Stat. Nonlinear Soft Matter. Phys. 81(4 Pt 2), 046106 (2010)
Blondel, V.D., et al.: Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008(10), P10008 (2008)
Bassett, D.S., et al.: Robust detection of dynamic community structure in networks. Chaos 23(1), 013142 (2013)
Guimerà, R., Amaral, L.A.: Cartography of complex networks: modules and universal roles. J. Stat. Mech. 2005(P02001), nihpa35573 (2005)
Hunter, J.D.: Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9(3), 90–95 (2007)
Tian, Y., et al.: Topographic organization of the human subcortex unveiled with functional connectivity gradients. Nat. Neurosci. 23(11), 1421–1432 (2020)
Kabbara, A., et al.: The dynamic functional core network of the human brain at rest. Sci. Rep. 7(1), 2936 (2017)
Betzel, R.F., et al.: Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage 127, 287–297 (2016)
Guimerà, R., Sales-Pardo, M., Amaral, L.A.: Classes of complex networks defined by role-to-role connectivity profiles. Nat. Phys. 3(1), 63–69 (2007)
Nakajima, M., Halassa, M.M.: Thalamic control of functional cortical connectivity. Curr. Opin. Neurobiol. 44, 127–131 (2017)
McFadyen, J., Dolan, R.J., Garrido, M.I.: The influence of subcortical shortcuts on disordered sensory and cognitive processing. Nat. Rev. Neurosci. 21(5), 264–276 (2020)
Pergola, G., et al.: The regulatory role of the human mediodorsal thalamus. Trends Cogn. Sci. 22(11), 1011–1025 (2018)
Achard, S., Bullmore, E.: Efficiency and cost of economical brain functional networks. PLOS Comput. Biol. 3(2), e17 (2007)
Sidibé, M., Paré, J.F., Smith, Y.: Nigral and pallidal inputs to functionally segregated thalamostriatal neurons in the centromedian/parafascicular intralaminar nuclear complex in monkey. J. Comp. Neurol. 447(3), 286–299 (2002)
Parent, A., Hazrati, L.-N.: Functional anatomy of the basal ganglia. II. The place of subthalamic nucleus and external pallidium in basal ganglia circuitry. Brain Res. Rev. 20(1), 128–154 (1995)
McFarland, N.R., Haber, S.N.: Thalamic relay nuclei of the basal ganglia form both reciprocal and nonreciprocal cortical connections, linking multiple frontal cortical areas. J. Neurosci. 22(18), 8117–8132 (2002)
Castro-Alamancos, M.A., Connors, B.W.: Thalamocortical synapses. Prog. Neurobiol. 51(6), 581–606 (1997)
Cole, M.W., Pathak, S., Schneider, W.: Identifying the brain’s most globally connected regions. Neuroimage 49(4), 3132–3148 (2010)
Guye, M., et al.: Graph theoretical analysis of structural and functional connectivity MRI in normal and pathological brain networks. Magn. Reson. Mater. Phys. Biol. Med. 23(5), 409–421 (2010)
de Pasquale, F., et al.: A cortical core for dynamic integration of functional networks in the resting human brain. Neuron 74(4), 753–764 (2012)
Kaiser, R.H., et al.: Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiat. 72(6), 603–611 (2015)
Mulders, P.C., et al.: Resting-state functional connectivity in major depressive disorder: a review. Neurosci. Biobehav. Rev. 56, 330–344 (2015)
Zhong, X., Pu, W., Yao, S.: Functional alterations of fronto-limbic circuit and default mode network systems in first-episode, drug-naïve patients with major depressive disorder: a meta-analysis of resting-state fMRI data. J. Affect. Disord. 206, 280–286 (2016)
Diagnostic and statistical manual of mental disorders: DSM-5, ed. A. American Psychiatric and D.S.M.T.F. American Psychiatric Association. 2013, Arlington, VA: American Psychiatric Association.
Jeste, D.V., Maglione, J.E.: Treating older adults with schizophrenia: challenges and opportunities. Schizophr. Bull. 39(5), 966–968 (2013)
Salman, M.S., et al.: Decreased cross-domain mutual information in schizophrenia from dynamic connectivity states. Front. Neurosci. 13, 873 (2019)
Perez-Costas, E., Melendez-Ferro, M., Roberts, R.C.: Basal ganglia pathology in schizophrenia: dopamine connections and anomalies. J. Neurochem. 113(2), 287–302 (2010)
Bernard, J.A., et al.: Patients with schizophrenia show aberrant patterns of basal ganglia activation: evidence from ALE meta-analysis. Neuroimage Clin. 14, 450–463 (2017)
Bassett, D.S., et al.: Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 28(37), 9239 (2008)
Liu, Y., et al.: Disrupted small-world networks in schizophrenia. Brain 131(4), 945–961 (2008)
Alexander-Bloch, A., et al.: The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage 59(4), 3889–3900 (2012)
Alexander-Bloch, A.F., et al.: Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front. Syst. Neurosci. 4, 147 (2010)
Pergola, G., et al.: The role of the thalamus in schizophrenia from a neuroimaging perspective. Neurosci. Biobehav. Rev. 54, 57–75 (2015)
Watis, L., et al.: Glutamatergic abnormalities of the thalamus in schizophrenia: a systematic review. J. Neural Transm. (Vienna) 115(3), 493–511 (2008)
Cieri, F., et al.: Brain entropy during aging through a free energy principle approach. Front. Hum. Neurosci. 15, 139 (2021)
Hutchison, R.M., et al.: Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80, 360–378 (2013)
Jones, D.T., et al.: Non-stationarity in the “resting brain’s” modular architecture. PLoS ONE 7(6), e39731 (2012)
Hansen, E.C., et al.: Functional connectivity dynamics: modeling the switching behavior of the resting state. Neuroimage 105, 525–535 (2015)
Thompson, W.H., Fransson, P.: A common framework for the problem of deriving estimates of dynamic functional brain connectivity. Neuroimage 172, 896–902 (2018)
Thompson, W.H., et al.: Simulations to benchmark time-varying connectivity methods for fMRI. PLoS Comput. Biol. 14(5), e1006196 (2018)
Karahanoglu, F.I., Van De Ville, D.: Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat. Commun. 6, 7751 (2015)
Hakimi, S.L.: On realizability of a set of integers as degrees of the vertices of a linear graph. I. J. Soc. Ind. Appl. Math. 10(3), 496–506 (1962)
Havel, V.: A remark on the existence of finite graphs. Casopis Pest. Mat. 80, 477–480 (1955)
Acknowledgements
Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hirsch, F., Wohlschlaeger, A. Graph analysis of nonlinear fMRI connectivity dynamics reveals distinct brain network configurations for integrative and segregated information processing. Nonlinear Dyn 108, 4287–4299 (2022). https://doi.org/10.1007/s11071-022-07413-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11071-022-07413-7