Abstract
The periodic transcription output is ubiquitously observed in an isogenic cell population. To understand mechanisms of cyclic behavior in transcription, we extend the gene activation process in the two-state model by assuming that the synthesis rate is periodic. We derive the analytical forms of the mean transcript level and the noise. The limits of them indicate that the mean level and the noise display periodic behaviors. Numerical examples strongly suggest that the transcription system with a periodic synthesis rate generates more noise than that with a constant rate but maintains transcription homeostasis in each period. It is also suggested that if the periodicity is not considered, the calculated noise may be greater than the real value.
Similar content being viewed by others
1 Introduction
Gene transcription in single cells is known to fluctuate stochastically in time and involves numerous successive steps [17, 22, 30]. The randomly switching of promoter between periods of active and inactive is thought to be the main source that leads to a large variability in mRNA levels. Two other functionally related processes to mRNA levels are synthesis and degradation. The synthesis rate is an important parameter in gene transcription, which may vary with time [28, 35, 46]. When intracellular environment varies, genes alter synthesis and/or degradation rates to readjust mRNA concentrations [28]. Such rates can be understood as upstream cellular drives that respond to changes of cellular environment [5].
Cyclic behavior is ubiquitous in transcription. Both in yeast and mammalian cells, a high proportion of genes express periodically [1]. Using high-density oligonucleotide microarrays and a cosine wave-fitting algorithm, Harmer et al. [11] identified clusters of circadian-regulated genes among more than 8000 genes. In many cells, there are some clock genes, whose transcriptional regulatory network plays an important role in generating cell-autonomous circadian rhythms [6, 19]. And the clock gene mechanism shows circadian oscillation of mRNA levels and time-related variations in the rate of change of clock gene expression [23]. Some transcriptional activators, such as BMAL1 and CLOCK, can regulate the period transcriptional repressor [19]. The mechanism of the circadian clock governs circadian transcript outputs of thousands of genes by affecting production, degradation of mRNAs and posttranscriptional regulation [21]. In addition, the light and temperature have the same effect on regulating gene expressions. The light and temperature cycles are the basic alternating environment in the world. Many organisms on Earth have adapted themselves to such alternating environment. Many fungi and yeast can exhibit periodic cellular activity to environmental stresses by using a dual sensing mechanism for temperature and light [26]. Recently, Zhuang et al. [45] uncovered that a circadian rhythm factor, BMAL1, is able to bind HBV genomes and increases viral promoter activity. In their experiments, synchronized differentiated HepaRG cells show a circadian cycling of Bmal1/Rev-\(Erb\alpha \) transcripts.
To understand molecular mechanisms of cyclic behavior in transcription, we describe the two-state model in detail by assuming the transcription rate to be periodic in Sect. 2. We then present the master equations and the differential equations. The transcriptional output and stochasticity of gene transcription have often been quantified by the mean, the noise and the noise strength [20, 22, 25, 27, 40]. For a random variable X, \(\mathrm{E}[X]\) denotes the mean value, and the noise and the noise strength are defined by
respectively, where \(\mathrm{Var}[X]\) denotes its variance. To characterize the stochasticity, we state the main mathematical results and present detailed proofs in Sect. 3. Our results show that the mean transcript level and the noise display periodic behaviors when the time is large. We perform numerical simulations to explore the contribution of the periodic mRNA synthesis rate on transcription in Sect. 4. We compare the mean level and the noise at different amplitudes of synthesis rates. The observations suggest that the increase in amplitude leads to large transcription noise, but maintains transcriptional output homeostasis in each period. With a synthesis rate like the cosine function, we derive the average level of transcripts and its average noise during a time interval. We find that the average noise may be larger than \(\eta ^2(t)\), even the maximal value. This demonstrates that the noise calculated from the experimental data is enlarged if the periodicity is not considered.
2 The model
2.1 The characterization of gene transcription
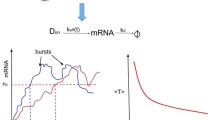
In this work, we continue to focus on a stochastic gene transcription model to investigate the dynamics of gene transcription and the fluctuation of transcripts. As usual, we employ the prevailing two-state model to characterize the transcription process, where mRNA transcripts are produced in burst in short active periods separated by long inactive periods. The main difference in this paper is that we consider the environment contribution to the transcription, where the environment changes periodically. Almost all living things in this world have developed their own intact circadian clock systems, which regulate their physiological mechanisms to answer daily changes in the environment. At the molecular level, mRNAs and proteins always oscillate in cycles of approximately 24-h to create the circadian rhythms. In fact, stimulated by cyclical environment, cells or organisms exhibit behavior with constant periodicity in many cases.
In this model, we hypothesize that the mRNA production rate \(\nu (t)\) is periodic with respect to t, that is,
where \(T>0\) is the period of \(\nu (t)\). To describe our model more clearly, we make some assumptions as follows:
-
(i)
The transcription system randomly rotates within the two functional states: the active (gene ON) state and the inactive (gene OFF) state.
-
(ii)
The durations in the ON and OFF states are independently and exponentially distributed with rates \(\gamma >0\) and \(\lambda >0\), respectively.
-
(iii)
Transcripts are produced with a synthesis rate \(\nu (t)\ge 0\) only when the gene is active and are turned over with a degradation rate \(\delta >0\) in each state.
-
(iv)
The synthesis rate \(\nu (t)\) has a constant period \(T>0\) and is bounded and integrable over any interval.
The OFF state is characterized by the lack of specific binding of the TF to the enhancer and no enzyme RNA polymerase elongating to the coding region. The entry of the ON state requires the TFs to bind at the enhancers to form a stable TF–DNA complex and the enzyme RNA polymerase to bind at the promoter. When the last engaged pol II leaves the coding region, the gene returns to the OFF state. Thus, Assumptions (i) and (ii) are basal and widely employed to model gene transcription. In mammalian cells, many intracellular behaviors are regulated by a circadian clock that generates intrinsic rhythms with a periodicity of approximately 24 hour [11]. Thus, Assumptions (iii) and (iv) should be considered to investigate gene expression.
As usual, we calculate the mean and the noise of transcript number. Let M(t) denote the transcript number of a gene of interest in single cells at time t. In single cells, M(t) is a natural number which varies over a very large region. In a population of isogenic cells, one expects to count how many transcripts produced per cell. That value is usually given by the first moment of M(t), that is,
To determine how the numbers of transcripts measured in individual cells deviate from the mean, we compute the noise
Clearly, the noise is completely determined by the first moment \(\hbox {E}[M(t)]\) and the second moment \(\hbox {E}[M^2(t)]\). From (3) and (4), both the mean and the noise are real-time values. It is difficult to derive these values using experimental data obtained at one time. With the help of approach named smFISH (single-molecule fluorescent in situ hybridization), mRNA abundance and transcriptional activity in yeast [42], mRNA copies in bacteria [9, 32] and in mammalian cells [30, 33] were measured. Usually, the transcript levels and the noises derived by using these data are close to the average transcript level and the average noise, which are defined by
where the second moment is
The two values in (5) describe the average level of transcripts during the time interval [0, t] and the corresponding noise.
2.2 The master equations
For any given time \(t\ge 0\), we let I(t) specify the promoter state, with \(I(t)=0\) when the promoter is in OFF state, and \(I(t)=1\) in ON state. Then, the transcription state can be fully quantified by the following joint probabilities
By using the Kolmogorov forward equations, we calculate the time evolutions of these probabilities (6) and (7), which give the master equations of our model. Suppose that the promoter resides at ON state with m copies of transcripts at time \(t+h\) for an infinitesimal time increment \(h>0\). Then, the basic Assumptions (i)–(iii) imply that, by discarding the events with transition probabilities of second or higher order of h, one of the four state transition events in Table 1 must occur during the time interval \((t, t+h)\). Adding the four probabilities listed in Table 1 gives
By dividing the resulted equality by h and then letting \(h\rightarrow 0\), we obtain
Next, we suppose that the promoter resides at OFF state with m copies of mRNA molecules at time \(t+h\). Since the transcription is closed when the promoter is inactivated, then there is no transcript being produced during \((t,t+h)\). By a similar discussion, we have
From Eq. (10), we obtain
By adding the joint probabilities \(P_0(m,t), P_1(m,t)\) in (6) and (7) when m takes all natural numbers, we derive two probabilities
The random switches of promoter between the two states can be considered as an alternating renewal process.
Stochastic gene transcription with a periodic synthesis rate. The promoter is activated by binding TFs at the enhancers forming a stable TF–DNA complex and binding RNA polymerase at the TATA box. RNA polymerases move along the template at the encoding region and synthesize RNA molecules with a periodic production rate \(\nu (t)\). RNA molecules are turned over with a constant rate \(\delta \)
2.3 The differential equations
We assume that the transcription starts from the gene OFF state and only consider the newly transcribed mRNA, which give the initial condition as
By adding (9) and (11) in m, we obtain a closed system of \(P_0(t)\) and \(P_1(t)\), that is,
and the initial values are \(P_0(0)=1\) and \(P_1(0)=0\). Solving (14) with the initial condition gives the following lemma.
Lemma 2.1
If the durations that the promoter resides at OFF and ON states are exponentially distributed with rates \(\lambda \) and \(\gamma \), and the initial condition (13) holds, then
It is easy to find that both \(P_0(t)\) and \(P_1(t)\) approach their constant limits
when time t tends to infinity, and \(P_1(t)\) is increasing over the time interval \((0,\infty )\). The two values \(P_0(t)\) and \(P_1(t)\) represent the probabilities that the cell resides OFF and ON states at time t. On the other hand, they also tell what percentages of cells reside at these two states at time t in a population of isogenic cells.
By definition, the mean number of mRNA molecules is
where P(m, t) is the probability that there exist exactly m transcripts in the cell at time t and is defined as
By differentiating (16) with respect to t and substituting (9) and (11) into the result, we derive
From the initial condition (13), we know \(m(0)=0\).
To calculate the second moment of transcripts, we need to define two mean levels
which shows that
The second moment of M(t) is
Differentiating (19) and with the assistance of (9) and (11) again, we obtain its time evolution as
The initial condition for \(\mu (t)\) is \(\mu (0)=0\).
Solving the two differential Eqs. (17) and (20) with their initial conditions, we will derive the analytical expressions of m(t) and \(\mu (t)\), and then, we can give the noise and the noise strength by their definitions (4).
3 Results
3.1 The mean transcript level
The external environment, such as light and temperature, and cellular environment, such as metabolism and resource supplies, always vary periodically, making mRNA and protein molecules also fluctuate periodically. In the section, we investigate the behavior of gene transcription which is regulated by the periodic changes in environment by assuming that the production rate \(\nu (t)\) is periodic over \((0,\infty )\). The following theorem tells us how many transcripts per cell one expects to count at time t in a population of isogenic cells and the average transcript number in one period when time t tends to infinity.
Theorem 3.1
Assume that the transcription of a gene obeys the model described in Fig. 1, and the mRNA synthesis rate \(\nu (t)\) is periodic and bounded. Under the initial condition (13), the expected value \(m(t)=\mathrm{E}[M(t)]\) of its mRNA copy number M(t) is given by
Let \(\langle m(t)\rangle _T\) be the expected value of m(t) over time interval \([t,t+T]\), that is,
Then, when time t tends to infinity, we have
Proof
The analytical expression of m(t) is obtained by solving the linear differential Eq. (17) with initial condition \(m(0)=0\). For any periodic function \(\nu (t)\), if it is integrable, we can derive the mean transcript level at time t by calculating (21).
When calculating the second moment of transcript number M(t), we need the analytical expressions of \(m_0(t)\) and \(m_1(t)\). From (16), the expression of m(t) can also be derived by summing \(m_0(t)\) and \(m_1(t)\). Multiplying (9) and (11) by m and summing up the products lead to
From the initial condition (13), when the transcription starts at time \(t=0\), the initial values of \(m_0(t)\) and \(m_1(t)\) are
We solve the two differential Eqs. (24) and (25) with their initial values and derive
Summing \(m_0(t)\) and \(m_1(t)\) together gives the analytical expression of m(t). The expected value of m(t) over \([t,t+T]\) can be obtained by substituting (21) into (22), that is,
By changing the order of integration, the integral is rewritten as
To derive (23), we let \(t\rightarrow \infty \) in \(\langle m(t)\rangle _T\). Since \(\nu (t)\) is bounded and integrable, and \(\lim _{t\rightarrow \infty }e^{-(\lambda +\gamma )t}=0\), by using L’Hôpital’s rule, we get
Note that \(P_1(t)\) has been given in Lemma 2.1. Then, the limit of the first integral is
We decompose the third integral into
When time t tends to infinity,
We only need to calculate
which equals
Since \(0<P_1(t+T)-P_1(t)\rightarrow 0\) as \(t\rightarrow \infty \) and \(\nu (t)\) is bounded, by applying L’Hôpital’s rule, we derive above limit. Now, we have obtained (23) and completed the proof. \(\square \)
In fact, from (23) it is easy to find that \(\int _0^T\nu (\tau )\mathrm{d}\tau /T\) is the average production rate over one period. Although many genes transcribe periodically, the average transcript numbers over one period always maintain homeostasis, no matter the synthesis rate is periodic or constant, or rewritten as
Since \(\nu (t)\) is periodic, m(t) has no limit as \(t\rightarrow \infty \). But the next theorem tells us that the mean level m(t) will tend to a periodic function with period T when \(t\rightarrow \infty \).
Theorem 3.2
Suppose that \(\nu (t)\) has positive period T. Then, there exists a unique periodic function
such that
Proof
We first prove that \(m^*(t)\) is periodic on \([0,\infty )\). From its expression, it is easy to find that
and thus, \(m^*(t)\) is periodic and continuous on \((0,\infty )\).
Next, we prove the limit (28) holds. Taking limits to \(m(t)-m^*(t)\) as \(t\rightarrow \infty \), we derive that
The limit holds since
Substituting \(m^*(t)\) into above limit, we have
We completed the proof. \(\square \)
Proposition 3.1
Let \(\langle m(t)\rangle \) be the expected value of transcripts over time interval [0, t], that is,
Then, we have
Proof
By definition, \(\langle m(t)\rangle \) is the average of the mean m(t) over time interval [0, t]. Taking limits to \(\langle m(t)\rangle \), we have
The last equality holds since \(m(t)-m^*(t)\rightarrow 0\) as \(t\rightarrow \infty \). For any \(t\ge T\), there is an integer \(k\ge 1\) such that \(kT\le t<(k+1)T\). Then,
Since \(m^*(t)\) is positive and periodic, we have
This completes the proof. \(\square \)
Lemma 3.1
Under the same condition of Theorem 3.1, when time t tends to infinity, \(m_1(t)\) tends to a periodic function \(m^*_1(t)\), where
3.2 The noise of transcripts
In this subsection, we will give the analytical formula of the noise function in integrals. Because of its technical complexity, the biological implication of the dynamical noise formula will not be discussed in detail. The limit of the noise as \(t\rightarrow \infty \) gives the “stationary” noise, which is also a periodic function.
Theorem 3.3
Under the same condition of Theorem 3.1, the second moment of the mRNA copy number M(t) is
Let \(\langle \mu (t)\rangle _T\) be the expected value of \(\mu (t)\) over the time interval \([t,t+T]\), that is,
Then, taking limits to \(\langle \mu (t)\rangle _T\) with respect to t, we have
Proof
We have given the time evolution of the second moment \(\mu (t)\) as shown in (20). Solving this differential equation with initial condition \(\mu (0)=0\), we can derive the analytical expression (31). Integrating \(\mu (t)\) over \([t,t+T]\) and dividing by T give \(\langle \mu (t)\rangle _T\), that is,
For simplicity, we use
to denote the integrand of the above integral. By the expression, we find that f(t) tends to a periodic function \(f^*(t)\), where
In fact
Theorem 3.2 and Lemma 3.1 have shown \(m(t)-m^*(t)\rightarrow 0\) and \(m_1(t)-m^*_1(t)\rightarrow 0\) when \(t\rightarrow \infty \). In Lemma 2.1, we have proved \(P_1(t)-P^*_1(t)\rightarrow 0\). Thus, f(t) goes to the periodic function \(f^*(t)\).
We reverse the order of integration in \(\langle \mu (t)\rangle _T\) to get
Now, we can calculate the limit \(\lim _{t\rightarrow \infty }\langle \mu (t)\rangle _T\) when t goes to infinity. In fact, the limit of the last two integrals in \(\langle \mu (t)\rangle _T\) is
Since \(\lim _{t\rightarrow \infty }[f(t)-f(t+T)]=0\), the first limit is
The integral \(\int _0^T f(\tau )e^{2\delta (\tau -T)}\mathrm{d}\tau \) is bounded, and the second limit is
Then,
We completed the proof. \(\square \)
Theorem 3.4
Under the same condition of Theorem 3.3, there exists a unique periodic function \(\mu ^*(t)\) such that
where \(\mu ^*(t)\) is defined as
with \(f^*(t)=2\nu (t)m^*_1(t)+\nu (t)P^*_1+\delta m^*(t)\).
Proof
By using a similar discussion as that in the proof of Theorem 3.2, we can prove that \(\mu ^*(t)\) is periodic and continuous on \([0,\infty )\). In fact,
We only need to show the limit (34) holds. It follows from the expressions of that
Taking limits to \(\mu ^*(t)-\mu (t)\) and noticing that
we see
In the proof of Theorem 3.3, we have shown that
By applying L’Hôpital’s rule, we derive the limit \(\lim _{t\rightarrow \infty }[\mu ^*(t)-\mu (t)]=0\). \(\square \)
Proposition 3.2
Let \(\langle \mu (t)\rangle \) be the average value of the second moment \(\mu (t)\) on the time interval (0, t], that is,
Then, when time t tends to infinity, we have
Proof
To get (37), we take limits to \(\langle \mu (t)\rangle \) with respect to t, that is,
Since \(\mu (t)-\mu ^*(t)\rightarrow 0\) when \(t\rightarrow \infty \), we get
For any \(t\ge T\), there exists an integer \(k\ge 1\) such that \(kT\le t\le (k+1)T\). Then,
Notice that \(\mu ^*(t)\) is positive and periodic. Taking limits in the above inequalities produces
This completes the proof. \(\square \)
Now, since the average transcript level m(t) and its second moment \(\mu (t)\) have been given, we can give the analytical expressions of noises. There are two different noises, which are defined as
The first noise (38) represents the fluctuation of transcripts at time t, which is a real-time value. From Theorems 3.2 and 3.4, we find \(\eta ^2(t)\) also approaches a periodic function when time t tends to infinity. The second one (39) represents the fluctuation of transcripts for all time on [0, t]. From the two Propositions 3.1 and 3.2, we find \(\langle \eta ^2(t)\rangle \) approaches a constant.
4 An example and simulations
4.1 The meal transcription level and the noise
Expressions of many mammalian genes have circadian clocks [39]. Using oligonucleotide arrays representing 12, 488 genes, Storch et al [34] identified 575 genes in liver and 462 genes in heart with circadian expression patterns. From the viewpoint that reaction rates will change with temperature or light, we present a general method to build a periodic gene transcription model. Genes are scored as circadian-regulated, if they have a greater correlation with a cosine test [11]. Johansson et al. [15] proposed to use the partial least square regression to identify genes with periodic fluctuations in expression levels coupled with the cell cycle in the budding yeast, where they used sine and cosine curves for fitting the observed expression profile for each gene. Kim et al. [18] used the Fourier series to approximate periodic biological phenomena. Increasing evidences suggest that periodic genes can be expressed as linear combinations of sine and cosine functions [2, 7, 8]. For this purpose, we consider a set of cosinusoid temperature variations with the period \(T=24\) hour [21, 38], which determines the transcription rate as
where \(\nu _0\) is the inherent transcription rate at some temperature, and the amplitude A is determined by the temperature difference between day and night. In the following simulations, we choose the inherent transcription rate \(\nu _0=1.89\) min\(^{-1}\) as suggested in [33]. To make sure that \(\nu (t)\) is positive, the amplitude A must be smaller than \(\nu _0\). Other data are determined by using smFISH to extract the kinetics of Oct4 [33], suggesting that gene activation rate and inactivation rate and the degradation rate for mature mRNA are
The mean transcript levels with different synthesis rates during fourteen days. When the synthesis rate \(\nu (t)\equiv \nu _0\) is a constant, the transcript level increases sharply and reaches an equilibrium state after one period. When \(\nu (t)\) is periodic with 24 hour, the average transcript level tends to a periodic function, which is also periodic with 24 hour. And the amplitude of the mean level is proportional to the amplitude of \(\nu (t)\). For the case \(A=90\), the mean transcript level m(t), as shown by the blue curve, is almost periodic after one period, but its average value on time interval [0, t], as shown by the red curve, tends to a steady value
When the synthesis rate \(\nu (t)\equiv \nu _0\) is a constant, the average transcript number approaches a steady state [13, 29, 31, 40], as shown by the black curve in Fig. 2. Even in the case where the degradation process is not instantaneous but takes some time, the time delay cannot lead to the occurrence of any oscillations [24]. But when \(\nu (t)\) is periodic, the simulation in Fig. 2 shows that the transcript level tends to a periodic function and oscillates around the black curve. Figure 2 also shows that the amplitude of the mean level increases with the amplitude of transcription rate. If the synthesis rate \(\nu (t)\) is periodic and its amplitude is \(A=90\), the mean transcript level is shown as the blue curve, which is almost periodic after one period, but its average value over the interval [0, t] approaches a constant, as shown by the red curve. And the limit of the average value on [0, t] is the same as the limit of m(t) with \(\nu (t)\equiv \nu _0\).
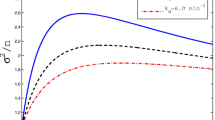
The noise of mRNA molecules. When \(\nu (t)\) is a constant, the noise \(\eta ^2(t)\) approaches its limit value (the black curve) after two periods. When \(\nu (t)\) is periodic, the noise \(\eta ^2(t)\) performs a periodic behavior after two periods. Same as the mean level m(t), the amplitude of the noise increases with the amplitude of \(\nu (t)\). When the time t goes to infinity, the average noise for transcriptional output over [0, t] approaches a steady value, which is greater than the limit of \(\eta ^2(t)\) with \(\nu (t)\equiv \nu _0\), as shown by the red curve
For the transcription system with a periodic rate \(\nu (t)\) as defined in (40), the mean transcript level is
whose average value over one period [0, T] is
When \(\nu (t)\equiv \nu _0\), the noise of transcripts is derived and discussed in several transcription models [4, 24, 31, 40]. In these models, the noise approaches the limit value. The behavior of \(\eta ^2(t)\) is shown by the black curve, which is almost a constant after two periods. In fact, for this case, the noise \(\eta ^2(t)\) has an exact limit, that is,
When \(\nu (t)\) is periodic, we have proved that both the first and second moments of transcripts tend to periodic functions in Theorem 3.2 and Theorem 3.4, respectively, and thus, the noise also tends to a periodic function. In Fig. 3, the simulation shows that \(\eta ^2(t)\) exhibits a periodic behavior and oscillates around the black curve after two periods and also shows the amplitude of \(\eta ^2(t)\) increases with the amplitude of \(\nu (t)\). When the amplitude \(A=90\) and the time \(t>2T\), the noise \(\eta ^2(t)\) varies over a narrow range [0.12, 0.24], shown by the blue curve. But we find that the average noise \(\langle \eta ^2(t)\rangle \) of transcripts over [0, t] tends to a constant value, that is,
which is greater than the maximal value of \(\eta ^2(t)\). By a simple calculation, we get
where
denotes the extra noise involved by the periodic behavior of the mRNA synthesis rate. From the expression of \(\Delta \), we find that \(\Delta \ge 0\), which indicates that the transcription system with a periodic synthesis rate always produces more noise than that with a constant rate.
According to the discussion above, the noise of transcripts may be magnified. Suppose that samples of experimental data come from different times. Then, the calculated values of the mean transcript level and the noise are close to \(m_T\) and \(\eta ^2_T\), respectively. If the synthesis rate \(\nu (t)\) is periodic but the periodicity is not considered in analyzing the transcription behavior, then \(\eta ^2_T\) may be greater than \(\eta ^2(t)\).
4.2 The amplitude and the delay time
With the development of real-time monitoring technique in single cells, a large amount of data has been produced. Numerous high-quality statistical approaches and algorithms are presented to estimate rhythmic parameters in large datasets. For instance, the JTK_ CYCLE is one of algorithms, which can accurately measure the period, phase and amplitude of cycling transcripts in large datasets [12]. Using these statistical approaches and algorithms, Zhang et al. [43] and Takahashi [37] detected a circadian gene expression atlas and a transcriptional architecture circadian clock in mammals, respectively.
With the help of the JTK_CYCLE algorithm, the oscillation period T and the absolute amplitude \(A_{m^*}\) of transcript abundance can be estimated from the experimental data. Then, using (40) and (43), we can obtain the amplitude and the approximate expression of \(\nu (t)\), which are essential for downstream analyses.
From (42), the amplitude of \(m^*(t)\) is
Since the mean degradation rate coefficient \(\delta \) is related to the average half-life by \(\tau _{1/2}=\ln 2/\delta \), the absolute amplitude of molecules deviating from the mean level \(m_T\) is
and the relative amplitude is
From (44) and (45), we find that the half-life has opposite effects on them. The long half-life enhances the absolute amplitude of transcript abundance, but reduces the relative amplitude, which is consistent with the result of previous study [21].
In experiments, it is found that the abundance of molecules has a later phase than the production rate if the molecule is rhythmically produced [21]. Let \(\tau \) be the delay time between the mean transcription level \(m^*(t)\) and the transcription rate \(\nu (t)\). For the transcription model that we present above, we can derive the delay time from (17), (40) and (42), that is,
which only depends on the period T and the degradation rate \(\delta \). It is clear that \(\tau \) has an upper bound T/4. For the example we give above, the delay time \(\tau \) is about 4.12 hour, as shown in Fig. 4.
5 Conclusion and discussion
Gene transcription in single cells is a complex and stochastic process, making the transcript copy number fluctuate in cell population. Highly variable mRNA distributions are the result of randomly switching between periods of active and inactive gene expression, and the birth and death process for the accumulation of mRNA molecules [14, 16, 36, 40,41,42, 44]. In addition, further sources of fluctuation, the noisy signals from heterogeneous environments are commonly invoked to explain the observed single-cell variability in mRNA numbers. To respond to the changes of the environment, cells generate diverse signal transduction mechanisms and transmit signals to the cell interior, resulting in changes in the expression of genes and the activity of enzymes by regulating the transcription activity [46], the degradation [10] or the switching between periods of active and inactive states [31].
Almost all light-sensitive organisms from bacteria to humans have a biological timekeeping mechanism to make our physiological mechanisms adapt daily changes in the environment, making cyclic behaviors be ubiquitous in transcription, which have important consequences for human health [3]. Therefore, it is important to understand molecular mechanisms of such behaviors. To delineate the contribution of periodic environmental signals to cell-to-cell variations in transcription, we studied the gene transcription behavior using the two-state model by assuming that the mRNA synthesis rate is periodic.
Our results showed that the periodicity of synthesis rate has a pronounced global role in affecting transcriptional output. It drove the mRNA molecules to be produced in a cyclical manner. Furthermore, the noise is proved to be periodic. In our simulation example, we consider circadian-regulated genes and assume that the synthesis rate is a cosine function. The simulation example verifies the periodicity of transcriptional output and its noise. As expected, the mean transcription level oscillates around the average value of transcript number over one period. Unexpectedly, the average noise may be larger than the mean noise. The simulation shows that the average noise is greater than the maximal value of the mean noise. Thus, the periodicity should be considered in investigating the stochastic behavior of transcription of circadian-regulated genes. If not, the fluctuation of the mRNA level calculated by experimental data may be amplified compared with its real value.
References
Breeden, L.L.: Periodic transcription: a cycle within a cycle. Curr. Biol. 13, 31–38 (2003)
Chen, J., Chang, K.C.: Discovering statistically significant periodic gene expression. Int. Stat. Rev. 76(2), 228–246 (2008)
Curtis, A.M., Fitzgerald, G.A.: Central and peripheral clocks in cardiovascular and metabolic function. Ann. Med. 38, 552–559 (2006)
Dar, R.D., Razooky, B.S., Weinberger, L.S., Cox, C.D., Simpson, M.L.: The low noise limit in gene expression. PLoS ONE 10(10), e0140969 (2015)
Dattani, J., Barahona, M.: Stochastic models of gene transcription with upstream drives: exact solution and sample path characterization. J. R. Soc. Interface 14, 20160833 (2016)
Erion, R., King, A.N., Wu, G., Hogenesch, J.B., Sehgal, A.: Neural clocks and neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. eLife 5, e13552 (2016)
Forger, D.B., Peskin, C.S.: A detailed predictive model of the mammalian circadian clock. PNAS 100, 14806–14811 (2003)
Forger, D.B., Peskin, C.S.: Model based conjectures on mammalian clock controversies. J. Theor. Biol. 230, 533–539 (2004)
Golding, I., Paulsson, J., Zawilski, S.M., Cox, E.C.: Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005)
Haimovich, G., Medina, D.A., Causse, S.Z., Garber, M., Millán-Zambrano, G., Barkai, O., Chávez, S., Pérez-Ortín, J.E., Darzacq, X., Choder, M.: Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000–1011 (2013)
Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., Kay, S.A.: Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 (2000)
Hughes, M.E., Hogenesch, J.B., Kornacker, K.: JTK\_CYCLE: an efficient non-parametric algorithm for detecting rhythmic components in genome-scale datasets. J. Biol. Rhythms 25, 372–380 (2010)
Jiao, F., Ren, J., Yu, J.: Analytical formula and dynamic profile of mRNA distribution. Discrete Contin. Dyn. Syst. B 25, 241–257 (2020)
Jiao, F., Tang, M., Yu, J.: Distribution modes and their corresponding parameter regions in stochastic gene transcription. SIAM J. Appl. Math. 75(6), 2396–2420 (2015)
Johansson, D., Lindgren, P., Berglund, A.: A multivariate approach applied to microarray data for identification of genes with cell cycle-coupled transcription. Bioinformatics 19, 467–473 (2003)
Kærn, M., Elston, T.C., Blake, W.J., Collins, J.J.: Stochasticity in gene expression: from theories to phenotypes. Nature 6, 451–464 (2005)
Kaufmann, B.B., van Oudenaarden, A.: Stochastic gene expression: from single molecules to the proteome. Curr. Opin. Genet. Dev. 17, 107–112 (2007)
Kim, B.R., Zhang, L., Berg, A., Fan, J., Wu, R.: A computational approach to the functional clustering of periodic gene-expression profiles. Genetics 18, 821–834 (2008)
Krishnaiah, S.Y., Wu, G., Altman, B.J., et al.: Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 25, 961–974 (2017)
Lin, G., Yu, J., Zhou, Z., Sun, Q., Jiao, F.: Fluctuations of mRNA distributions in multiple pathway activated transcription. Discrete Contin. Dyn. Syst. B 24, 1543–1568 (2019)
Lück, S., Thurley, K., Thaben, P.F., Westermark, P.O.: Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 9, 741–751 (2014)
Maheshri, N., O’Shea, E.K.: Living with noisy genes: how cells function reliably with inherent variability in gene expression. Ann. Rev. Biophys. Biomol. Struct. 36, 413–434 (2007)
Mazzoccoli, G., Francavilla, M., Giuliani, F., Aucella, F., Vinciguerra, M., Pazienza, V., Piepoli, V., Benegiamo, G., Liu, S., Cai, Y.: Clock gene expression in mouse kidney and testis: analysis of periodical and dynamical patterns. J. Biol. Regul. Homeost. Agents 26(2), 303–311 (2012)
Miȩkisz, J., Poleszczuk, J., Bodnar, M., Foryś, U.: Stochastic models of gene expression with delayed degradation. Bull. Math. Biol. 73, 2231–2247 (2011)
Munsky, B., Neuert, G., van Oudenaarden, A.: Using gene expression noise to understand gene regulation. Science 336, 183–187 (2012)
Okamoto, S., Furuya, K., Nozaki, S., Aoki, K., Niki, H.: Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot. Cell 12, 1235–1243 (2013)
Paulsson, J.: Summing up the noise in gene networks. Nature 427, 415–418 (2004)
Pérez-Ortín, J.E., Alepuz, P.M., Moreno, J.: Genomics and gene transcription kinetics in yeast. Trends Genet. 23, 250–257 (2007)
Pérez-Ortín, J.E., Medina, D.A., Chávez, S., Moreno, J.: What do you mean by transcription rate? The conceptual difference between nascent transcription rate and mRNA synthesis rate is essential for the proper understanding of transcriptomic analyses. BioEssays 35, 1056–1062 (2013)
Raj, A., Peskin, C.S., Tranchina, D., Vargas, D.Y., Tyagi, S.: Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, 1707–1719 (2006)
Ren, J., Jiao, F., Sun, Q., Tang, M., Yu, J.: The dynamics of gene transcription in random environments. Discrete Contin. Dyn. Syst. Ser. B 23, 3167–3194 (2018)
Skinner, S.O., Sepúlveda, L.A., Xu, H., Golding, I.: Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 6, 1100–1113 (2013)
Skinner, S.O., Xu, H., Nagarkar-Jaiswal, S., Freire, P.R., Zwaka, T.P., Golding, I.: Single-cell analysis of transcription kinetics across the cell cycle. Life 5, e12175 (2016)
Storch, K.F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F.C., Wong, W.H., Weitz, C.J.: Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83 (2002)
Sun, Q., Jiao, F., Lin, G., Yu, J., Tang, M.: The nonlinear dynamics and fluctuations of mRNA levels in cell cycle coupled transcription. PLoS Comput. Biol. 15(4), e1007017 (2019)
Sun, Q., Tang, M., Yu, J.: Modulation of gene transcription noise by competing transcription factors. J. Math. Biol. 64, 469–494 (2012)
Takahashi, J.S.: Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017)
Takeuchi, T., Hinohara, T., Kurosawa, G., Uchid, K.: A temperature-compensated model for circadian rhythms that can be entrained by temperature cycles. J. Theor. Biol. 246, 195–204 (2007)
Vollmers, C., Gill, S., DiTacchio, L., Pulivarthy, S.R., Le, H.D., Panda, S.: Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. PNAS 106, 21453–21458 (2009)
Yu, J., Sun, Q., Tang, M.: The nonlinear dynamics and fluctuations of mRNA levels in cross-talking pathway activated transcription. J. Theor. Biol. 363, 223–234 (2014)
Yu, J., Xiao, J., Ren, X., Lao, K., Xie, X.S.: Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006)
Zenklusen, D., Larson, D.R., Singer, R.H.: Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008)
Zhang, R., Lahens, N.F., Ballance, H.I., Hughes, M.E., Hogenesch, J.B.: A circadian gene expression atlas in mammals: implications for biology and medicine. PNAS 111, 16219–16224 (2014)
Zhu, C., Han, G., Jiao, F.: Dynamical regulation of mRNA distribution by cross-talking signaling pathways. Complexity 2020, 6402703 (2020)
Zhuang, X., Forde, D., Tsukuda, S., et al.: Circadian control of hepatitis B virus replication. Nat. Commun. 12, 1658 (2021)
Zopf, C.J., Quinn, K., Zeidman, J., Maheshri, N.: Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput. Biol. 9(7), e1003161 (2013)
Funding
This study was funded by the National Natural Science Foundation of China (11631005, 11871174) and the Science and Technology Program of Guangzhou (201707010337).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and design of the study. Material preparation, data collection and analysis were performed by Qiwen Sun and Feng Jiao. The first draft of the manuscript was written by Jianshe Yu, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Q., Jiao, F. & Yu, J. The dynamics of gene transcription with a periodic synthesis rate. Nonlinear Dyn 104, 4477–4492 (2021). https://doi.org/10.1007/s11071-021-06569-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11071-021-06569-y