Abstract
Purpose
To investigate the potential of radiomics applied to static clinical PET data using the tracer O-(2-[18F]fluoroethyl)-l-tyrosine (FET) to differentiate treatment-related changes (TRC) from tumor progression (TP) in patients with gliomas.
Patients and Methods
One hundred fifty-one (151) patients with histologically confirmed gliomas and post-therapeutic progressive MRI findings according to the response assessment in neuro-oncology criteria underwent a dynamic amino acid PET scan using the tracer O-(2-[18F]fluoroethyl)-l-tyrosine (FET). Thereof, 124 patients were investigated on a stand-alone PET scanner (data used for model development and validation), and 27 patients on a hybrid PET/MRI scanner (data used for model testing). Mean and maximum tumor to brain ratios (TBRmean, TBRmax) were calculated using the PET data from 20 to 40 min after tracer injection. Logistic regression models were evaluated for the FET PET parameters TBRmean, TBRmax, and for radiomics features of the tumor areas as well as combinations thereof to differentiate between TP and TRC. The best performing models in the validation dataset were finally applied to the test dataset. The diagnostic performance was assessed by receiver operating characteristic analysis.
Results
Thirty-seven patients (25%) were diagnosed with TRC, and 114 (75%) with TP. The logistic regression model comprising the conventional FET PET parameters TBRmean and TBRmax resulted in an AUC of 0.78 in both the validation (sensitivity, 64%; specificity, 80%) and the test dataset (sensitivity, 64%; specificity, 80%). The model combining the conventional FET PET parameters and two radiomics features yielded the best diagnostic performance in the validation dataset (AUC, 0.92; sensitivity, 91%; specificity, 80%) and demonstrated its generalizability in the independent test dataset (AUC, 0.85; sensitivity, 81%; specificity, 70%).
Conclusion
The developed radiomics classifier allows the differentiation between TRC and TP in pretreated gliomas based on routinely acquired static FET PET scans with a high diagnostic accuracy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the follow-up of glioma patients, treatment-related changes often cannot be reliably differentiated from tumor progression by structural magnetic resonance imaging (MRI) alone. Yet a false diagnosis will either result in the continuation of an ineffective treatment or a premature termination of an effective one, both negatively impacting patients’ prognosis. To improve the differentiation of treatment-related changes and tumor progression, advanced MRI techniques such as perfusion-weighted imaging as well as MR spectroscopy are under investigation, yielding accuracies of about 80% for this clinically challenging and highly important differential diagnosis [1].
Besides advanced MRI, amino acid positron emission tomography (PET) has demonstrated its potential to differentiate treatment-related changes from tumor progression in initial studies [2,3,4]. Previous studies using the amino acid PET tracer O-(2-[18F]fluoroethyl)-l-tyrosine (FET) showed that the combination of static and dynamic parameters discriminates treatment-related changes from tumor progression in recurrent gliomas with an accuracy of up to 90% [5,6,7]. These studies, however, require dynamic FET PET parameters based on a 40–50 min PET scan that is more time consuming in clinical routine than a static scan from 20 to 40 min post injection. Other studies demonstrated an improved diagnostic accuracy by combination of FET PET with advanced MRI methods using hybrid PET/MRI [8, 9].
Despite these recent advances, a method to improve the diagnostic performance of FET PET without the need for a time-consuming and expensive dynamic acquisition or additional, dedicated MRI scans would be of clinical relevance.
In recent years, methods based on artificial intelligence and machine learning have become increasingly important and found their way into medical image analysis. Several methods from this field are under investigation also in brain tumor patients and promise to improve diagnosis by extracting additional imaging features from routinely acquired imaging data. These features are usually not accessible through conventional image analysis and can be used to generate prognostic or predictive mathematical models. This methodology is also referred to as radiomics [10, 11].
FET PET radiomics has already demonstrated its potential in neurooncology for the prediction of the isocitrate dehydrogenase genotype [12], the diagnosis of pseudoprogression [13, 14], the differentiation of treatment-related changes from recurrent brain metastases after radiosurgery [15, 16], or the prediction of the BRAF mutational status in patients with melanoma brain metastases [17].
The goal of our study was to investigate the potential of FET PET radiomics for the differentiation between treatment-related changes and tumor progression in patients with glioma based on routinely acquired static FET PET when added to clinically established FET PET parameters.
Patients and methods
Patients
The patient group was partly included in a previous study concerning the diagnostic performance of perfusion-weighted MRI and dynamic FET PET for the differentiation of treatment-related changes from glioma progression [18].
One hundred and fifty-one patients (n = 54 females, n = 97 males; median age, 52.3 years; age range 20.4–78.0 years) with histologically confirmed WHO grade II–IV glioma according to the 2016 WHO Classification of Tumors of the Central Nervous System [19] were included in this retrospective study [mostly WHO grade IV glioblastoma, IDH-wildtype (n = 71); WHO grade III anaplastic astrocytoma, IDH-mutant (n = 20); WHO grade II astrocytoma, IDH-mutant (n = 14)]. All patients presented with post-therapeutic MRI findings suspicious for tumor progression according to the Response Assessment in Neuro-Oncology (RANO) criteria and were hence investigated using FET PET. All patients included here showed an increased FET uptake in the area of the primary lesion. Complete patient characteristics are provided in Table 1.
Diagnosis of treatment-related changes and tumor progression
Diagnosis was based on histopathology in 46 patients (30%), and on clinicoradiological follow-up in 105 patients (70%). For histopathologic diagnosis, tissue samples were obtained by resection or biopsy and analyzed as described previously [5].
For WHO grade II gliomas, both the clinical and the radiological situation had to be stable or improved for at least 12 months without change in therapy to exclude tumor progression [20]. For WHO grade III–IV gliomas, the diagnosis treatment-related changes required at least 6 months of stable or improved clinical and radiological condition [21], as well as no change in tumor treatment. Tumor progression was diagnosed if lesions continued to increase in size on at least two subsequent MRI scans according to the RANO criteria, accompanied by a deterioration in performance status, or if a patient died of glioma, whichever occurred first. Of note, the applied classification criteria are in accordance with previous studies [5, 22, 23].
FET PET imaging
The amino acid FET was produced and applied as described previously [24]. According to international guidelines for brain tumor imaging, all patients fasted for at least 4 h before the PET measurement [25].
All patients underwent a dynamic PET scan from 0 to 50 min post injection of 3 MBq of FET per kg of body weight. 124 patients were examined on a stand-alone PET scanner (ECAT EXACT HR+, Siemens Healthcare, Erlangen, Germany) in 3D mode, and 27 patients on a high-resolution 3 T hybrid PET/MRI scanner (BrainPET, Siemens Healthcare, Erlangen, Germany). The BrainPET is a compact cylinder that fits into the bore of the Magnetom Trio MR scanner [26, 27].
As described before [28], iterative reconstruction parameters were: 16 subsets, six iterations using the OSEM algorithm for the ECAT HR+ PET scanner and two subsets, and 32 iterations using the OP-OSEM algorithm for the BrainPET. Data were corrected for random, scattered coincidences, dead time, and motion for both systems. Attenuation correction for the ECAT HR+ PET was based on a transmission scan. For the BrainPET, a template-based approach was used [26]. The reconstructed dynamic data sets consisted of 16 time frames (5 × 1 min; 5 × 3 min; 6 × 5 min) for both scanners. To optimize comparability of the results related to the influence of the two different PET scanners, reconstruction parameters, and post-processing steps, a 2.5 mm 3D Gaussian filter was applied to the BrainPET data before further processing. In previous phantom experiments with spheres of different sizes that simulated lesions, this filter kernel demonstrated the best comparability of PET data obtained from the ECAT HR+ PET and the BrainPET scanner [29].
Evaluation of FET PET parameters
The FET uptake was expressed as standardized uptake value (SUV) by dividing the radioactivity in the tissue (kBq/ml) by the radioactivity injected per gram of body weight.
Semi-automated segmentation of the suspected brain lesion was performed in the summed PET images from 20 to 40 min post-injection using the Pmod Biomedical Image Quantification Software (Version 3.806, PMOD Technologies, Zurich, Switzerland). For assessment of the FET uptake in healthy brain tissue, a region-of-interest was positioned in the semioval center of the unaffected hemisphere contralateral to the tumor, involving grey and white matter [25]. As described before [28], tumor segmentation was performed using a 2-dimensional auto-contouring process in the transversal slice containing the voxel with the maximum tracer uptake using a tumor-to-brain ratio (TBR) of 1.6 or more. In a previous study, this threshold has been shown to best separate between vital tumor and healthy brain parenchyma in FET PET [30].
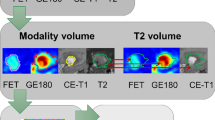
Maximum and mean TBRs (TBRmax, TBRmean) were calculated by dividing the maximum or mean SUV of the tumor by the mean SUV of healthy brain tissue. The segmentations were validated by an experienced, board-certified specialist in neuroradiology with broad experience in nuclear medicine. Figure 1 illustrates the segmented lesions in representative patients.
Representative FET PET images of patients with treatment-related changes (top) and glioma progression (bottom). The segmented lesions are highlighted in red in the right column. Visually, obvious differences in FET uptake between patients with treatment-related changes and tumor progression could not be identified
Image preprocessing and radiomics feature extraction
The group of patients scanned on the stand-alone PET scanner was divided into a training and a validation dataset in a ratio of 3/1 with an equal ratio of tumor progression to treatment-related changes. The group of patients scanned on the 3 T hybrid PET/MR scanner was used for model testing.
Feature extraction was performed by the RadiomiX toolbox (supported by Oncoradiomics, Liège, Belgium) [31] implemented in Matlab 2017a (MathWorks, Natick, MA, USA), including International Biomarker Standardization Initiative (IBSI)-compliant [32] radiomic features as well as others. A total of 221 features were extracted. No spatial resampling was performed. Absolute intensity resampling was performed using a fixed bin width of 0.1 according to current recommendations [33].
The extracted features consisted of five main groups: (1) fractal features (2) first order statistics, (3) shape and size, (4) texture descriptors including gray level co-occurrence (GLCM), gray level run-length (GLRLM), and gray level size-zone texture matrices (GLSZM), and (5) features from groups 1, 3 and 4 after wavelet decomposition of the original image.
The definitions and detailed feature descriptions are provided elsewhere [10]. Detailed mathematical definitions of the features are available in the RadiomiX toolbox manual.
Feature selection
Using large number of features on a limited number of patients for model calculation may result in data overfitting. Overfitting is a methodological mistake in which a generated model corresponds too closely or even matches the analyzed dataset. This results in a perfect classification accuracy on the dataset that has been used for training but renders the model too specialized to classify new or additional imaging data or reliably predict future observations. To lower the risk of overfitting, the most important features must be identified in a process called feature selection before model generation. [34]
Feature selection was performed using the RadiomiX toolbox. First, the machine learning module eliminates features with (near) zero variance and an inter-feature correlation of 0.9 or more to remove redundancies within the feature set. A threshold of 0.9 is generally considered high enough to eliminate high correlation from the dataset. Second, a stepwise forward feature selection using stratified cross-validation with logistic regression is used to further reduce the number of radiomics features, i.e., it stops adding features if the inclusion of the next feature does not add more than 0.005 to the average cross-validation AUC.
Model generation and validation
Logistic regression models were generated on the training dataset using the ‘tidyverse/ggplot2’ packages in R (version 4.0.5, R Studio, Inc., Boston, MA, USA). Logistic regression models were fitted separately for the conventional FET PET parameters TBRmean and TBRmax, as well as for the selected radiomics features and a combination of conventional and radiomics features. Finally, the models were applied to the holdout validation dataset that was not part of model generation.
Model testing
The best performing models were applied to the test dataset acquired on the BrainPET scanner. Since the test dataset was not involved in the process of model training and validation and was acquired on a different PET scanner, it represents an independent dataset to evaluate the robustness and generalizability of the model. The radiomics workflow is illustrated in Fig. 2.
Statistical evaluation
Descriptive statistics are provided as mean and standard deviation or as median and range. The Mann–Whitney-U test was used for intergroup comparison. The diagnostic performance of the PET parameters, the machine learning models and combinations thereof were assessed by receiver operating characteristic (ROC) analysis. The decision cutoff was considered optimal when the product of paired values for sensitivity and specificity reached its maximum. Fisher’s exact test for 2 × 2 contingency tables was used for statistical evaluation of the parameters. P-values of less than 0.05 were considered statistically significant. Statistical analyses were performed using SPSS (SPSS Statistics 24, IBM, New York, USA) and Microsoft Excel (Excel:Mac 2020, Version 16.53, Microsoft, Redmond, WA, USA).
Results
Treatment-related changes and tumor progression
Of the 124 patients examined on the stand-alone PET scanner, 31 (25%) were diagnosed with treatment-related changes and 93 (75%) with tumor progression. The test dataset consisted of 27 patients examined on the BrainPET scanner. Thereof, six (22%) were diagnosed with treatment-related changes and 21 (78%) with tumor progression.
Group comparison of FET PET parameters
TBRmean was significantly higher for patients with tumor progression compared to patients with treatment-related changes (mean TBRmean ± standard deviation, 2.1 ± 0.3 vs. 1.9 ± 0.3; p < 0.001). TBRmax was also significantly higher for patients with tumor progression compared to patients with treatment-related changes (mean TBRmax ± standard deviation, 3.7 ± 0.9 vs. 2.8 ± 0.7; p < 0.001).
Performance of machine learning models in the training and validation dataset
The two most important radiomics features according to the feature selection were Informational Measure of Correlation 2 calculated from the GLCM, and Intensity Non-Uniformity Normalized from the GLSZM.
The logistic regression model based on the conventional PET parameters resulted in an AUC of 0.78 (95% confidence interval, 0.68–0.88; sensitivity, 64%; specificity, 80%) in the validation dataset. The logistic regression model using only radiomics features resulted in an AUC of 0.90 (95% confidence interval, 0.79–1.00; sensitivity, 87%; specificity, 80%) in the validation dataset. The logistic regression model combining FET PET parameters and radiomics features resulted in an AUC of 0.92 (95% confidence interval, 0.82–1.00; sensitivity, 91%; specificity, 80%) in the validation dataset. Further details on the model performances in the validation and test dataset are summarized in Table 2 and Fig. 3.
Receiver operating characteristic curves of the validation (left) and the test dataset (right) for a model comprising the static FET PET parameters TBRmean and TBRmax (top row), a model comprising the two radiomics features Informational Measure of Correlation 2 calculated from the grey level co-occurrence matrix, and Intensity Non-Uniformity Normalized from the grey level size zone matrix (middle row), and a model using the combination thereof (bottom row)
Performance of radiomics models in the test dataset
The model based on PET parameters resulted in an AUC of 0.78 (95% confidence interval, 0.67–0.88; sensitivity, 66%; specificity, 80%) in the test dataset.
The model based solely on radiomics features resulted in an AUC of 0.85 (95% confidence interval, 0.77–0.94; sensitivity, 73%; specificity, 80%) in the test dataset. The model combining FET PET parameters and radiomics features resulted in an AUC of 0.85 (95% confidence interval, 0.77–0.94; sensitivity, 81%; specificity, 70%). Further details on the model performances in the test dataset are summarized in Table 2 and Fig. 3.
Discussion
The main finding of our study is that a machine learning model based on static FET PET radiomics features differentiates treatment-related changes from tumor progression in patients with gliomas with a high diagnostic accuracy, i.e., an AUC of 0.85, and outperforms conventional FET PET analysis (AUC, 0.78). Interestingly, combining both FET PET radiomics features with conventional FET PET parameters further improved the overall diagnostic performance, especially the sensitivity of the model. Further, the developed radiomics model was evaluated in a small, but independent test dataset acquired with a different PET scanner demonstrating its reliability and robustness regardless of the used scanners and imaging parameters. Since the developed model is based on routinely acquired FET PET scans and can be applied fully automated on a conventional computer in a few minutes, the approach seems feasible for clinical implementation.
Several studies already investigated the potential of FET PET for the differentiation of treatment-related changes from tumor progression in glioma patients [7, 20, 35, 36]. Although these studies demonstrate high diagnostic accuracies, the number of patients in these studies was either low, and/or only a small fraction of patients were diagnosed with treatment-related changes. A more recent study by Maurer and colleagues [5] using partially overlapping patients investigated the value of static and dynamic FET PET parameters for the differentiation of treatment-related changes from glioma progression. This retrospective analysis of 127 patients with WHO grade II-IV gliomas yielded a diagnostic accuracy of 81% (sensitivity, 86%; specificity, 67%) by combining TBRmax and the dynamic FET PET parameter slope. In our study, the combination of static FET PET parameters TBRmean and TBRmax resulted in a comparable diagnostic performance (AUC, 0.78; sensitivity, 64%; specificity, 80%) in the validation dataset. Nevertheless, the use of FET PET radiomics parameters alone already outperformed the conventional FET PET parameters with an AUC of 0.90 (sensitivity, 87%; specificity, 80%). Combining radiomics and conventional FET PET parameters further increased the diagnostic performance in the validation dataset (AUC, 0.92; sensitivity, 91%; specificity, 80%) and demonstrated its generalizability in the external test dataset (AUC, 0.85; sensitivity, 81%; specificity, 70%).
Our results concerning FET PET are also comparable with a recent study of our group investigating the value of combining perfusion-weighted MRI with dynamic FET PET [37]. In that study, accuracy of PWI, which can be performed easily during routine conventional MR scanning, was poor in differentiating treatment-related changes and tumor progression (accuracy, 63%). However, the high positive predictive value of PWI (100%) allowed a correct diagnosis of treatment-related changes in 42% of the patients. In the remaining patients, PWI was nondiagnostic, but FET PET still achieved an accuracy of 78% leading to the recommendation of a sequential use of perfusion-weighted MRI and dynamic FET PET in clinical practice. In this context, the developed radiomics classifier in our study may achieve a higher diagnostic performance based on 20 min static FET PET images. Since the analysis can be performed fully automated on a conventional computer in a few minutes, this combination appears promising in terms of a clinical translation.
Over the past years, the value of machine learning techniques and FET PET radiomics for the diagnosis of treatment-related changes such as pseudoprogression in patients with glioma [13, 14, 38] or radiation injury in patients with brain metastases [16] has been demonstrated. Interestingly, in these studies, different patterns of tracer uptake could already visually be distinguished. Patients with pseudoprogression or radiation injuries showed a more homogenous uptake of FET compared to a more heterogenous uptake of FET in patients with tumor progression.
In our study, different patterns of FET uptake between patients with treatment-related changes and glioma progression could not be identified by visual evaluation (Fig. 1). This might be due to the more inhomogeneous group of patients that included a broad range of glioma subtypes and treatment regimens. Nevertheless, we identified the two textural features Informational Measure of Correlation 2 calculated from the GLCM, and Intensity Non-Uniformity Normalized from the GLSZM as being discriminative between treatment-related changes and glioma progression. Both textural features describe differences in tumor heterogeneity not accessible by means of human perception. Even though visually accessible differences are desirable for a better presentation of the results, the real benefit and concept of radiomics becomes more apparent if this is not the case—”images are more than pictures, they are data” [39].
Besides a visual interpretation of radiomics features, further efforts are needed for a deeper understanding of the biological meaning of features and machine learning models. This might be essential for a potential clinical translation and acceptance of radiomics in clinical routine. Hence, the correlation of radiomics features with local tissue samples including extensive neuropathological work-up is necessary in future studies.
Although promising, our results must be further validated in a larger group of patients from multiple institutions. Albeit the number of patients in our study is larger than in other studies investigating PET radiomics for the differentiation of treatment-related changes and tumor progression, the generally low number of patients available in neuro-oncology remains a limitation. Nonetheless, our model has shown its value in an external test dataset without extensive preprocessing, acquired on a different PET scanner, so further model evaluation in other centers is warranted and feasible.
Another limitation of our study might be the heterogenous composition of patients in terms of glioma subtypes and treatment regimens and the relatively low amount of histopathological validation of the diagnosis. Further, the group of patients is likely biased towards more challenging cases as only patients with equivocal MRI findings and remaining therapeutic options usually undergo FET PET scans. However, this dataset is representative of a clinical situation and further underlines the value of the model as it does not require an extensive preselection of patients.
Additionally, the developed machine learning model is based on FET PET alone and does not include structural or advanced MRI. Future studies should hence address the potential additional value of a combined FET PET/MRI radiomics analysis, also considering advanced MRI methods such as PWI or MR spectroscopy.
Conclusion
The results from our study suggest that the developed radiomics model is of clinical value for the differentiation between treatment-related changes and tumor progression in patients with gliomas regardless of tumor type or pretreatment. The radiomics model is based on routinely acquired static 20 min FET PET scans facilitating the translation into clinical routine. Especially in combination with other clinical parameters, the developed radiomics model might have an additional diagnostic value once translated into clinical routine.
Data availability
All data supporting the findings presented in this manuscript are available upon request directly to the corresponding author, PL. These data are not part of public domain or database as they are part of the patient protected medical record and public sharing would compromise the privacy of the research participants.
References
Anselmi M, Catalucci A, Felli V, Vellucci V, Di Sibio A, Gravina GL et al (2017) Diagnostic accuracy of proton magnetic resonance spectroscopy and perfusion-weighted imaging in brain gliomas follow-up: a single institutional experience. Neuroradiol J 30:240–252. https://doi.org/10.1177/1971400916688354
Cui M, Zorrilla-Veloz RI, Hu J, Guan B, Ma X (2021) Diagnostic accuracy of PET for differentiating true glioma progression from post treatment-related changes: a systematic review and meta-analysiS. Front Neurol 12:671867. https://doi.org/10.3389/fneur.2021.671867
Heiss WD (2017) Positron emission tomography imaging in gliomas: applications in clinical diagnosis, for assessment of prognosis and of treatment effects, and for detection of recurrences. Eur J Neurol 24:1255-e70. https://doi.org/10.1111/ene.13385
Langen KJ, Galldiks N, Hattingen E, Shah NJ (2017) Advances in neuro-oncology imaging. Nat Rev Neurol 13:279–89. https://doi.org/10.1038/nrneurol.2017.44
Maurer GD, Brucker DP, Stoffels G, Filipski K, Filss CP, Mottaghy FM et al (2020) (18)F-FET PET imaging in differentiating glioma progression from treatment-related changes: a single-center experience. J Nucl Med 61:505–11. https://doi.org/10.2967/jnumed.119.234757
Pöpperl G, Kreth FW, Herms J, Koch W, Mehrkens JH, Gildehaus FJ et al (2006) Analysis of 18F-FET PET for grading of recurrent gliomas: is evaluation of uptake kinetics superior to standard methods? J Nucl Med 47:393–403
Galldiks N, Stoffels G, Filss C, Rapp M, Blau T, Tscherpel C et al (2015) The use of dynamic O-(2–18F-fluoroethyl)-l-tyrosine PET in the diagnosis of patients with progressive and recurrent glioma. Neuro Oncol 17:1293–300. https://doi.org/10.1093/neuonc/nov088
Pyka T, Hiob D, Preibisch C, Gempt J, Wiestler B, Schlegel J et al (2018) Diagnosis of glioma recurrence using multiparametric dynamic 18F-fluoroethyl-tyrosine PET-MRI. Eur J Radiol 103:32–7. https://doi.org/10.1016/j.ejrad.2018.04.003
Lohmeier J, Bohner G, Siebert E, Brenner W, Hamm B, Makowski MR (2019) Quantitative biparametric analysis of hybrid (18)F-FET PET/MR-neuroimaging for differentiation between treatment response and recurrent glioma. Sci Rep 9:14603. https://doi.org/10.1038/s41598-019-50182-4
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–62. https://doi.org/10.1038/nrclinonc.2017.141
van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B (2020) Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 11:91. https://doi.org/10.1186/s13244-020-00887-2
Lohmann P, Lerche C, Bauer EK, Steger J, Stoffels G, Blau T et al (2018) Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep 8:13328. https://doi.org/10.1038/s41598-018-31806-7
Lohmann P, Elahmadawy MA, Gutsche R, Werner JM, Bauer EK, Ceccon G et al (2020) FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers (Basel) 12:3835. https://doi.org/10.3390/cancers12123835
Kebir S, Khurshid Z, Gaertner FC, Essler M, Hattingen E, Fimmers R et al (2017) Unsupervised consensus cluster analysis of [18F]-fluoroethyl-L-tyrosine positron emission tomography identified textural features for the diagnosis of pseudoprogression in high-grade glioma. Oncotarget 8:8294–304. https://doi.org/10.18632/oncotarget.14166
Lohmann P, Stoffels G, Ceccon G, Rapp M, Sabel M, Filss CP et al (2017) Radiation injury vs. recurrent brain metastasis: combining textural feature radiomics analysis and standard parameters may increase (18)F-FET PET accuracy without dynamic scans. Eur Radiol 27:2916–2927. https://doi.org/10.1007/s00330-016-4638-2
Lohmann P, Kocher M, Ceccon G, Bauer EK, Stoffels G, Viswanathan S et al (2018) Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. Neuroimage Clin 20:537–42. https://doi.org/10.1016/j.nicl.2018.08.024
Meissner AK, Gutsche R, Galldiks N, Kocher M, Junger ST, Eich ML et al (2021) Radiomics for the non-invasive prediction of the BRAF mutation status in patients with melanoma brain metastases. Neuro Oncol. https://doi.org/10.1093/neuonc/noab294
Steidl E, Langen KJ, Hmeidan SA, Polomac N, Filss CP, Galldiks N et al (2021) Sequential implementation of DSC-MR perfusion and dynamic [(18)F]FET PET allows efficient differentiation of glioma progression from treatment-related changes. Eur J Nucl Med Mol Imaging 48:1956–65. https://doi.org/10.1007/s00259-020-05114-0
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–20. https://doi.org/10.1007/s00401-016-1545-1
Popperl G, Gotz C, Rachinger W, Gildehaus FJ, Tonn JC, Tatsch K (2004) Value of O-(2-[18F]fluoroethyl)-L-tyrosine PET for the diagnosis of recurrent glioma. Eur J Nucl Med Mol Imaging 31:1464–70. https://doi.org/10.1007/s00259-004-1590-1
Mehrkens JH, Popperl G, Rachinger W, Herms J, Seelos K, Tatsch K et al (2008) The positive predictive value of O-(2-[18F]fluoroethyl)-L-tyrosine (FET) PET in the diagnosis of a glioma recurrence after multimodal treatment. J Neurooncol 88:27–35. https://doi.org/10.1007/s11060-008-9526-4
Werner JM, Stoffels G, Lichtenstein T, Borggrefe J, Lohmann P, Ceccon G et al (2019) Differentiation of treatment-related changes from tumour progression: a direct comparison between dynamic FET PET and ADC values obtained from DWI MRI. Eur J Nucl Med Mol Imaging 46:1889–901. https://doi.org/10.1007/s00259-019-04384-7
Mihovilovic MI, Kertels O, Hanscheid H, Lohr M, Monoranu CM, Kleinlein I et al (2019) O-(2-((18)F)fluoroethyl)-L-tyrosine PET for the differentiation of tumour recurrence from late pseudoprogression in glioblastoma. J Neurol Neurosurg Psychiatry 90:238–239. https://doi.org/10.1136/jnnp-2017-317155
Hamacher K, Coenen HH (2002) Efficient routine production of the 18F-labelled amino acid O-2-18F fluoroethyl-L-tyrosine. Appl Radiat Isot 57:853–856. https://doi.org/10.1016/s0969-8043(02)00225-7
Law I, Albert NL, Arbizu J, Boellaard R, Drzezga A, Galldiks N et al (2019) Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [(18)F]FDG: version 1.0. Eur J Nucl Med Mol Imaging 46:540–557. https://doi.org/10.1007/s00259-018-4207-9
Herzog H, Langen KJ, Weirich C, Rota Kops E, Kaffanke J, Tellmann L et al (2011) High resolution BrainPET combined with simultaneous MRI. Nuklearmedizin 50:74–82. https://doi.org/10.3413/Nukmed-0347-10-09
Caldeira L (2019) The Julich experience with simultaneous 3T MR-BrainPET: methods and technology. IEEE Trans Radiat Plasma Med Sci 3:352–62
Bauer EK, Stoffels G, Blau T, Reifenberger G, Felsberg J, Werner JM et al (2020) Prediction of survival in patients with IDH-wildtype astrocytic gliomas using dynamic O-(2-[(18)F]-fluoroethyl)-L-tyrosine PET. Eur J Nucl Med Mol Imaging 47:1486–95. https://doi.org/10.1007/s00259-020-04695-0
Lohmann P, Herzog H, Rota Kops E, Stoffels G, Judov N, Filss C et al (2015) Dual-time-point O-(2-[(18)F]fluoroethyl)-L-tyrosine PET for grading of cerebral gliomas. Eur Radiol 25:3017–24. https://doi.org/10.1007/s00330-015-3691-6
Pauleit D, Floeth F, Hamacher K, Riemenschneider MJ, Reifenberger G, Muller HW et al (2005) O-(2-[18F]fluoroethyl)-L-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain 128:678–87. https://doi.org/10.1093/brain/awh399
Keek S, Sanduleanu S, Wesseling F, de Roest R, van den Brekel M, van der Heijden M et al (2020) Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri)tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS ONE 15:e0232639. https://doi.org/10.1371/journal.pone.0232639
Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–38. https://doi.org/10.1148/radiol.2020191145
Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R et al (2015) The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 5:11075. https://doi.org/10.1038/srep11075
Stegmayr C, Stoffels G, Filss C, Heinzel A, Lohmann P, Willuweit A et al (2021) Current trends in the use of O-(2-[(18)F]fluoroethyl)-L-tyrosine ([(18)F]FET) in neurooncology. Nucl Med Biol 92:78–84. https://doi.org/10.1016/j.nucmedbio.2020.02.006
Rachinger W, Goetz C, Popperl G, Gildehaus FJ, Kreth FW, Holtmannspotter M et al (2005) Positron emission tomography with O-(2-[18F]fluoroethyl)-l-tyrosine versus magnetic resonance imaging in the diagnosis of recurrent gliomas. Neurosurgery 57:505–511. https://doi.org/10.1227/01.neu.0000171642.49553.b0
Galldiks N, Dunkl V, Stoffels G, Hutterer M, Rapp M, Sabel M et al (2015) Diagnosis of pseudoprogression in patients with glioblastoma using O-(2-[18F]fluoroethyl)-L-tyrosine PET. Eur J Nucl Med Mol Imaging 42:685–95. https://doi.org/10.1007/s00259-014-2959-4
Steidl E, Langen KJ, Hmeidan SA, Polomac N, Filss CP, Galldiks N et al (2020) Sequential implementation of DSC-MR perfusion and dynamic [(18)F]FET PET allows efficient differentiation of glioma progression from treatment-related changes. Eur J Nucl Med Mol Imaging. https://doi.org/10.1007/s00259-020-05114-0
Kebir S, Schmidt T, Weber M, Lazaridis L, Galldiks N, Langen KJ et al (2020) A preliminary study on machine learning-based evaluation of static and dynamic FET-PET for the detection of pseudoprogression in patients with IDH-wildtype glioblastoma. Cancers (Basel) 12:3080. https://doi.org/10.3390/cancers12113080
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–77. https://doi.org/10.1148/radiol.2015151169
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project numbers 491111487 and 428090865/SPP 2177 (RG, NG, and PL).
Author information
Authors and Affiliations
Contributions
Study conception and design: MM, ES, EH, JPS, GDM, FM, AH, NG, K-JL, PL Data acquisition: MK, CPF, GS, ES, EH, JPS, GDM, AH, NG, K-JL. Data analysis and interpretation: MM, OW, RG, RTHL, MK, PL. Writing draft of the manuscript: MM, PL All authors commented on previous versions of the manuscript, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interest to disclose.
Ethical approval
The local ethics committees approved the retrospective analysis of the data (project number, SNO-8-2018). The study adheres to the standards established in the declaration of Helsinki.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Müller, M., Winz, O., Gutsche, R. et al. Static FET PET radiomics for the differentiation of treatment-related changes from glioma progression. J Neurooncol 159, 519–529 (2022). https://doi.org/10.1007/s11060-022-04089-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-022-04089-2