Abstract
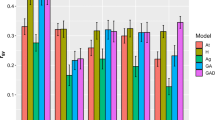
Genomic selection (GS) is poised to revolutionize eucalypt tree improvement by shortening breeding cycles and increasing selection intensities. This could be particularly valuable for alternative, non-mainstream Eucalyptus species that are still in the initial stages of breeding. Eucalyptus benthamii is important for its adaptation to frost-prone subtropical regions. In this work, we compared seven genomic prediction models, six Bayesian and one frequentist GBLUP (Genomic Best Linear Unbiased Prediction) with the conventional pedigree-based ABLUP approach. Models were evaluated for their ability to estimate heritabilities and predict wood quality traits (wood density, extractives, lignin, and carbohydrates content) and volume growth in 77 open-pollinated families of Eucalyptus benthamii. We also evaluated predictive abilities and heritabilities using variable numbers of SNP in the models. Heritabilities ranged from 0.09 (extractives content) using Bayesian Lasso (BL) to 0.55 (wood density) using ABLUP. Predictive abilities (PA) ranged from 0.12 (for volume using ABLUP) to 0.44 (for wood density using three Bayesian models). All seven genomic models performed similarly well and better than the pedigree model for all traits, except extractives content. Subsets of 5000–7000 SNPs yielded heritabilities and PAs nearly as large as using all 15,293 SNPs. However, a low-density SNP panel might not be economically and technically advantageous compared to the current high-density multi-species Eucalyptus EUCHIP60k. Our results support a positive outlook to implement GS to accelerate Eucalyptus benthamii breeding for adaptation to frost-prone regions.



Similar content being viewed by others
Data Archiving Statement
Genotypic (10.6084/m9.figshare.14806449) and phenotypic (10.6084/m9.figshare.14807916) data used in this study are available in the Figshare Digital Repository.
References
ABNT (2003) Wood—Determination of basic density. NBR 11941:2003. ABNT—Assoc Bras Normas Técnicas 6
Beaulieu J, Doerksen T, Clément S et al (2014a) Accuracy of genomic selection models in a large population of open-pollinated families in white spruce. Heredity (edinb) 113:343–352. https://doi.org/10.1038/hdy.2014.36
Beaulieu J, Doerksen TK, MacKay J, Rainville A, Bousquet J (2014b) Genomic selection accuracies within and between environments and small breeding groups in white spruce. BMC Genom 15:1048
Bhandari A, Bartholomé J, Cao-Hamadoun TV et al (2019) Selection of trait-specific markers and multi-environment models improve genomic predictive ability in rice. PLoS ONE 14:e0208871. https://doi.org/10.1371/JOURNAL.PONE.0208871
Butcher PA, Skinner AK, Gardiner CA (2005) Increased inbreeding and inter-species gene flow in remnant populations of the rare Eucalyptus benthamii. Conserv Genet 6:213–226. https://doi.org/10.1007/s10592-004-7830-x
Butnor JR, Johnsen KH, Anderson PH et al (2019) Growth, photosynthesis, and cold tolerance of Eucalyptus benthamii planted in the Piedmont of North Carolina. For Sci 65:59–67. https://doi.org/10.1093/forsci/fxy030
Chen Z-Q, Baison J, Pan J et al (2018) Accuracy of genomic selection for growth and wood quality traits in two control-pollinated progeny trials using exome capture as the genotyping platform in Norway spruce. BMC Genom 19:946. https://doi.org/10.1186/s12864-018-5256-y
da Costa RML, Estopa RA, Biernaski FA, Mori ES (2016) Predição de ganhos genéticos em progênies de Eucalyptus benthamii Maiden et Cambage por diferentes métodos de seleção. Sci for. https://doi.org/10.18671/scifor.v44n109.10
Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA (2010) The impact of genetic architecture on genome-wide evaluation methods. Genetics 185:1021–1031. https://doi.org/10.1534/genetics.110.116855
De Almeida Filho JE, Guimarães JFR, Fonsceca E, Silva F et al (2019) Genomic prediction of additive and non-additive effects using genetic markers and pedigrees. G3 Genes Genomes Genet 9:2739–2748. https://doi.org/10.1534/g3.119.201004
De Los Campos G, Naya H, Gianola D et al (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182:375–385. https://doi.org/10.1534/genetics.109.101501
De Moraes BFX, dos Santos RF, de Lima BM et al (2018) Genomic selection prediction models comparing sequence capture and SNP array genotyping methods. Mol Breed. https://doi.org/10.1007/s11032-018-0865-3
Denis M, Favreau B, Ueno S et al (2013) Genetic variation of wood chemical traits and association with underlying genes in Eucalyptus urophylla. Tree Genet Genomes 9:927–942. https://doi.org/10.1007/s11295-013-0606-z
Do C, Waples RS, Peel D et al (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214. https://doi.org/10.1111/1755-0998.12157
DoVale JC, Carvalho HF, Sabadin F, Fritsche-Neto R (2021) Reduction of genotyping marker density for genomic selection is not an affordable approach to long-term breeding in cross-pollinated crops. bioRxiv 2021.03.05.434084. https://doi.org/10.1101/2021.03.05.434084
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4:250–255. https://doi.org/10.3835/plantgenome2011.08.0024
Ferraz AG, Cruz CD, dos Santos GA et al (2020) Potential of a population of Eucalyptus benthamii based on growth and technological characteristics of wood. Euphytica 216:1–15. https://doi.org/10.1007/s10681-020-02628-4
Fonseca SM, Resende MDV, Alfenas AC et al (2010) Manual Prático de Melhoramento Genético do Eucalipto, 1st edn. Universidade Federal de Viçosa, Viçosa
Gao H, Su G, Janss L et al (2013) Model comparison on genomic predictions using high-density markers for different groups of bulls in the Nordic Holstein population. J Dairy Sci 96:4678–4687. https://doi.org/10.3168/jds.2012-6406
Geweke J (1992) Evaluating the accuracy of sampling-based approaches to calculating posterior moments. Bayesian Stat 4:641
Ghoreishifar SM, Moradi-Shahrbabak H, Fallahi MH et al (2020) Genomic measures of inbreeding coefficients and genome-wide scan for runs of homozygosity islands in Iranian river buffalo, Bubalus bubalis. BMC Genet. https://doi.org/10.1186/s12863-020-0824-y
Gianola D, van Kaam JBCHM (2008) Reproducing kernel hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178:2289–2303. https://doi.org/10.1534/genetics.107.084285
Gianola D, Fernando RL, Stella A (2006) Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173:1761–1776. https://doi.org/10.1534/genetics.105.049510
Gianola D, De Los Campos G, González-Recio O et al (2010) Statistical learning methods for genome-based analysis of quantitative traits. In: 9th world congress of genetics applied to livestock production, Leipzig, Germany, pp 1–6
Goldschmid O (1971) Ultraviolet spectra. In: Sarknanen K, Ludwig C (eds) Lignin: occurrence, formation, structure and reactions. Wiley, New York, pp 241–266
Gomide J, Demuner B (1986) Determination of lignin in woody material: modified Klason method. O Pap 47:36–38
González-Recio O, Gianola D, Rosa GJ et al (2009) Genome-assisted prediction of a quantitative trait measured in parents and progeny: application to food conversion rate in chickens. Genet Sel Evol 41:3. https://doi.org/10.1186/1297-9686-41-3
Gorjanc G, Cleveland MA, Houston RD, Hickey JM (2015) Potential of genotyping-by-sequencing for genomic selection in livestock populations. Genet Sel Evol 47:12. https://doi.org/10.1186/s12711-015-0102-z
Grattapaglia D, Kirst M (2008) Eucalyptus applied genomics: from gene sequences to breeding tools. New Phytol 179:911–929
Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genomes 7:241–255. https://doi.org/10.1007/s11295-010-0328-4
Grattapaglia D, Silva-Junior OB, Resende RT et al (2018) Quantitative genetics and genomics converge to accelerate forest tree breeding. Front Plant Sci 871:1–10. https://doi.org/10.3389/fpls.2018.01693
Grattapaglia D (2014) Breeding forest trees by genomic selection: current progress and theway forward. In: Genomics of plant genetic resources: volume 1. Managing, sequencing and mining genetic resources. Springer, Netherlands, pp 651–682
Habier D, Fernando RL, Kizilkaya K, Garrick DJ (2011) Extension of the Bayesian alphabet for genomic selection. BMC Bioinform 12:186. https://doi.org/10.1186/1471-2105-12-186
Hall KB, Stape J, Bullock BP et al (2019) A growth and yield model for Eucalyptus benthamii in the southeastern United States. For Sci. https://doi.org/10.1093/forsci/fxz061
Hall N, Brooker MIH (1973) Camden white gum, Eucalyptus benthamii Maiden et Cambage. Australian Government Pub. Service
Han L, Love K, Peace B et al (2020) Origin of planted Eucalyptus benthamii trees in Camden NSW: checking the effectiveness of circa situm conservation measures using molecular markers. Biodivers Conserv 29:1301–1322. https://doi.org/10.1007/s10531-020-01936-4
Hill WG, Weir BS (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78. https://doi.org/10.1016/0040-5809(88)90004-4
Ibá (2021) 2021—Ibá Annual Report, São Paulo
Isik F, Bartholomé J, Farjat A et al (2016) Genomic selection in maritime pine. Plant Sci 242:108–119. https://doi.org/10.1016/j.plantsci.2015.08.006
Isik F, Holland J, Maltecca C et al (2017) Genomic relationships and GBLUP. In: Genetic data analysis for plant and animal breeding. Springer, pp 311–354
Kainer D, Stone EA, Padovan A et al (2018) Accuracy of genomic prediction for foliar terpene traits in Eucalyptus polybractea. G3 Genes Genomes Genet 8:2573–2583. https://doi.org/10.1534/g3.118.200443
Kärkkäinen HP, Sillanpää MJ (2012) Back to basics for Bayesian model building in genomic selection. Genetics 191:969–987. https://doi.org/10.1534/genetics.112.139014
Kjaer E, Amaral W, Yanchuk A, Graudal L (2004) Strategies for conservation of forest genetic resources. Conservation of Eucalyptus benthamii: an endangered eucalypt species from eastern Australia. In: Forest genetic resources conservation and management: overview, concepts and some systematic approaches, 1st edn. Interntional Plant Genetic Resources Institute, Rome, pp 5–24
Klápště J, Suontama M, Telfer E et al (2017) Exploration of genetic architecture through sib-ship reconstruction in advanced breeding population of Eucalyptus nitens. PLoS ONE 12:e0185137. https://doi.org/10.1371/journal.pone.0185137
Lee SH, Clark S, Van Der Werf JHJ (2017) Estimation of genomic prediction accuracy from reference populations with varying degrees of relationship. PLoS ONE. https://doi.org/10.1371/journal.pone.0189775
Lenz PRN, Beaulieu J, Mansfield SD et al (2017) Factors affecting the accuracy of genomic selection for growth and wood quality traits in an advanced-breeding population of black spruce (Picea mariana). BMC Genom 18:335. https://doi.org/10.1186/s12864-017-3715-5
Lenz PRN, Nadeau S, Mottet M et al (2020) Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway spruce. Evol Appl 13:76–94. https://doi.org/10.1111/eva.12823
Lima BM, Cappa EP, Silva-Junior OB et al (2019) Quantitative genetic parameters for growth and wood properties in Eucalyptus “urograndis” hybrid using near-infrared phenotyping and genome-wide SNP-based relationships. PLoS ONE 14:1–24. https://doi.org/10.1371/journal.pone.0218747
Lin M, Arnold RJ, Li B, Yang M (2003) Selection of cold-tolerant eucalypts for Hunan Province. In: Turnbull JW (ed) Proceedings of Eucalypts in Asia—a symposium held in Zhanjiang, People’s Republic of China, 7–11 April 2003. ACIAR Proceedings. Australian Centre for International Agricultural Research, Zhanjiang, Guangdong, People’s Republic of China. Canberra, pp 107–116
Long N, Gianola D, Rosa GJM, Weigel KA (2011) Long-term impacts of genome-enabled selection. J Appl Genet 52:467–480. https://doi.org/10.1007/s13353-011-0053-1
Marroni F, Pinosio S, Zaina G et al (2011) Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet Genomes 7:1011–1023. https://doi.org/10.1007/s11295-011-0391-5
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mora AL, Garcia CH (2000) Eucalypt cultivation in Brazil. Sociedade Brasileira de Silvicultura, São Paulo
Mphahlele MM, Isik F, Mostert-O’Neill MM et al (2020) Expected benefits of genomic selection for growth and wood quality traits in Eucalyptus grandis. Tree Genet Genomes 16:49. https://doi.org/10.1007/s11295-020-01443-1
Müller BSF, Neves LG, de Almeida Filho JE et al (2017) Genomic prediction in contrast to a genome-wide association study in explaining heritable variation of complex growth traits in breeding populations of Eucalyptus. BMC Genom 18:1–17. https://doi.org/10.1186/s12864-017-3920-2
Muñoz PR, Resende MFR, Gezan SA et al (2014) Unraveling additive from nonadditive effects using genomic relationship matrices. Genetics 198:1759–1768. https://doi.org/10.1534/genetics.114.171322
Myburg AA, Potts BM, Marques CM et al (2007) Eucalypts. In: Kole C (ed) Forest trees, 1st edn. Springer, Berlin, pp 115–160
Namkoong G, Kang HC, Brouard JS (1988) Tree breeding: principles and strategies. Springer, New York
Ødegård J, Meuwissen THE (2012) Estimation of heritability from limited family data using genome-wide identity-by-descent sharing. Genet Sel Evol 44:16. https://doi.org/10.1186/1297-9686-44-16
Paludeto JGZ, Grattapaglia D, Estopa RA, Tambarussi EV (2021) Genomic relationship-based genetic parameters and prospects of genomic selection for growth and wood quality traits in Eucalyptus benthamii. Tree Genet Genomes 17:38. https://doi.org/10.1007/s11295-021-01516-9
Park T, Casella G (2008) The Bayesian Lasso. J Am Stat Assoc 103:681–686. https://doi.org/10.1198/016214508000000337
Pérez P, De Los Campos G (2014) Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495. https://doi.org/10.1534/genetics.114.164442
Pirraglia A, Gonzalez R, Saloni D et al (2012) Fuel properties and suitability of Eucalyptus benthamii and Eucalyptus macarthurii for torrefied wood and pellets. BioResources 7:217–235
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Ratcliffe B, Gamal El-Dien O, Klápště J et al (2015) A comparison of genomic selection models across time in interior spruce (Picea engelmannii × glauca) using unordered SNP imputation methods. Heredity (Edinb) 115:547–555. https://doi.org/10.1038/hdy.2015.57
Ratcliffe B, El-Dien OG, Cappa EP et al (2017) Single-step BLUP with varying genotyping effort in open-pollinated Picea glauca. G3 Genes Genomes Genet 7:935–942. https://doi.org/10.1534/g3.116.037895
Resende MFR Jr, Muñoz P, Resende MDV et al (2012a) Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190:1503–1510. https://doi.org/10.1534/genetics.111.137026
Resende MDV, Resende MFR, Sansaloni CP et al (2012b) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194:116–128. https://doi.org/10.1111/j.1469-8137.2011.04038.x
Resende RT, Resende MDV, Silva FF et al (2017) Assessing the expected response to genomic selection of individuals and families in Eucalyptus breeding with an additive-dominant model. Heredity (Edinb) 119:245–255. https://doi.org/10.1038/hdy.2017.37
Silva-Junior OB, Grattapaglia D (2015) Genome-wide patterns of recombination, linkage disequilibrium and nucleotide diversity from pooled resequencing and single nucleotide polymorphism genotyping unlock the evolutionary history of Eucalyptus grandis. New Phytol 208:830–845. https://doi.org/10.1111/nph.13505
Silva-Junior OB, Faria DA, Grattapaglia D (2015) A flexible multi-species genome-wide 60K SNP chip developed from pooled resequencing of 240 Eucalyptus tree genomes across 12 species. New Phytol 206:1527–1540. https://doi.org/10.1111/nph.13322
Skinner A (2003) The effects of tree isolation on the genetic diversity and seed production of Camden White Gum (Eucalyptus benthamii Maiden et Cambage). In: Centre for Plant Biodiversity Research. http://www.cpbr.gov.au/cpbr/summer-scholarship/2002-projects/skinner-alison-report.html. Accessed 12 May 2020
Suontama M, Klápště J, Telfer E et al (2019) Efficiency of genomic prediction across two Eucalyptus nitens seed orchards with different selection histories. Heredity (Edinb) 122:370–379. https://doi.org/10.1038/s41437-018-0119-5
Tambarussi EV, Pereira FB, da Silva PHM et al (2018) Are tree breeders properly predicting genetic gain? A case study involving Corymbia species. Euphytica 214:1–11. https://doi.org/10.1007/s10681-018-2229-9
Tan B, Grattapaglia D, Martins GS et al (2017) Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol. https://doi.org/10.1186/s12870-017-1059-6
Tan B, Grattapaglia D, Wu HX, Ingvarsson PK (2018) Genomic relationships reveal significant dominance effects for growth in hybrid Eucalyptus. Plant Sci 267:84–93. https://doi.org/10.1016/j.plantsci.2017.11.011
TAPPI TA of the P and P (2000) Tappi T280 pm-99 standard—acetone extractives of wood and pulp. TAPPI Press
Thavamanikumar S, McManus LJ, Tibbits JFG, Bossinger G (2011) The significance of single nucleotide polymorphisms (SNPs) in Eucalyptus globulus breeding programs. Aust For 74:23–29. https://doi.org/10.1080/00049158.2011.10676342
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423. https://doi.org/10.3168/jds.2007-0980
Wallis AFA, Wearne RH, Wright PJ (1996) Analytical characteristics of plantation eucalypt woods relating to kraft pulp yields. Appita J 49:427–432
Wang X, Miao J, Chang T et al (2019) Evaluation of GBLUP, BayesB and elastic net for genomic prediction in Chinese simmental beef cattle. PLoS ONE. https://doi.org/10.1371/journal.pone.0210442
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756. https://doi.org/10.1111/j.1755-0998.2007.02061.x
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262. https://doi.org/10.1111/j.1752-4571.2009.00104.x
White TL, Adams WT, Neale DB (2007) Forest genetics. CABI, Wallingford
Xavier A (2019) Efficient estimation of marker effects in plant breeding. G3 Genes Genomes Genet 9:3855–3866. https://doi.org/10.1534/g3.119.400728
Zhang Q, Calus MP, Guldbrandtsen B et al (2015) Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. https://doi.org/10.1186/s12863-015-0227-7
Acknowledgements
João Gabriel Zanon Paludeto received scholarships from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Evandro V. Tambarussi and Dario Grattapaglia were supported by research productivity fellowships granted by CNPq (Brazilian National Council for Scientific and Technological Development).
Funding
This work was partially supported by PRONEX-FAP-DF Grant 2009/00106-8 ‘NEXTREE’, CNPq (Brazilian Council for Scientific and Technological Development) Grant 400663/2012-0 and EMBRAPA Grant 03.11.01.007.00.00 to DG that allowed the development and validation of the multi-species EuCHIP60K used in this work.
Author information
Authors and Affiliations
Contributions
RAE Methodology, Formal analysis, Investigation, Resources, Data curation, Funding acquisition, Writing—Original Draft. JGZP Software, Formal analysis, Investigation, Data curation, Writing—Original Draft. BMSF Methodology, Formal analysis, Software, Investigation, Data curation, Writing—Original Draft. RAO Conceptualization, Methodology. CFA Formal analysis, Data curation, Writing—Review and Editing. MDVR Conceptualization, Methodology, EVT Visualization, Writing—Review and Editing. DG Conceptualization, Methodology, Investigation, Resources, Writing—Final Review and Editing, Supervision, Project administration.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Consent to participate
Not applicable
Consent for publication
Not applicable
Ethical approval
Not applicable
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Estopa, R.A., Paludeto, J.G.Z., Müller, B.S.F. et al. Genomic prediction of growth and wood quality traits in Eucalyptus benthamii using different genomic models and variable SNP genotyping density. New Forests 54, 343–362 (2023). https://doi.org/10.1007/s11056-022-09924-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-022-09924-y