Abstract
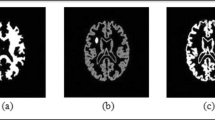
The spread of lung tumors and their changes occur dynamically, so precise segmentation of the images obtained is necessary. In this study, an extended region growth algorithm was performed on CT lung tumor images to examine accurate tumor margin and area. At first, a new threshold was implemented in MATLAB software by defining a larger target region around the primary tumor. Then, nearby points were settled in an array and then these points were updated based on tumor growth to set the fresh tumor margins. By the algorithm, furthest distance from the center of color intensity point of the primary tumorous area was selected to grow the region. Afterwards, fresh tumor border was determined by interpolation between these refreshed points through drawing lines from the tumor region center. The edge correction was then applied and the obtained new region was attached to the main region to reach a segmented tumor exterior. This technique improved the tumor recognition by 96% accuracy. In the inclusive algorithm, the conformance percentage had a positive impact on the achievement of the threshold and the renewal of the relative amount by 13% over the accuracy score. Also compared to the basilar algorithm, at least 12% of the percent differences in conformity were found to segment the tumor region in lung CT images. The derived dice similarity coefficients were close to each other for both the basilar and inclusive algorithms by 0.79±0.05 and 0.88±0.04, correspondingly. The p-value of these dice coefficients was less than 0.08 resulting from the paired Student’s t-test between two algorithms. The combination of methods such as machine learning is intended to improve segmentation accuracy for different types of nodule and tumor CT images.

















Similar content being viewed by others
Data availability
All data collected from the database used by DIR lab are available on the Winship Cancer Institute website (https://www.dir-lab.com).
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study. All data required to support the results and conclusions of the study have been provided here with the submission.
Abbreviations
- CT:
-
Computed tomography
- ROI:
-
Region of interest
- RA:
-
Relative amount
- DIR:
-
Deformable image registration
References
Cao M, Wang S, Wei L, Rai L, Li D, Yu H, Shao D (2019) Segmentation of immunohistochemical image of lung neuroendocrine tumor based on double layer watershed. Multimed Tools Appl 78:9193–9215. https://doi.org/10.1007/s11042-018-6431-5
Barros Netto SM, Silva AC, Cardoso de Paiva A, Nunes RA, Gattass M (2017) Unsupervised detection of density changes through principal component analysis for lung lesion classification. Multimed Tools Appl 76:18929–18954. https://doi.org/10.1007/s11042-017-4414-6
Yadav DP, Jalal AS, Goyal A, Mishra A, Uprety K, Guragai N (2023) COVID-19 radiograph prognosis using a deep CResNeXt network. Multimed Tools Appl 82:36479–36505. https://doi.org/10.1007/s11042-023-14960-7
Hosseini MP, Soltanian-Zadeh H, Akhlaghpoor S (2012) Detection and severity scoring of chronic obstructive pulmonary disease using volumetric analysis of lung CT images. Iran J Radiol. 9(1):22–7. https://doi.org/10.5812/iranjradiol.6759
Soleymanpour E, Pourreza HR, Ansaripour E, Yazdi MS (2011) Fully automatic lung segmentation and rib suppression methods to improve nodule detection in chest radiographs. J Med Signals Sens 1(3):191–199 n/a
Varshini PR, Baskar S, Alagappan S (2012) An improved adaptive border marching algorithm for inclusion of juxtapleural nodule in lung segmentation of CT-images. In: Venugopal KR, Patnaik LM (eds) Wireless Networks and Computational Intelligence. ICIP 2012. Communications in Computer and Information Science, vol 292. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-31686-9_27
Suji RJ, Bhadauria SS, Godfrey W, Dhar J (2023) On using a particle image velocimetry based approach for candidate nodule detection. Multimed Tools Appl 82:22871–22888. https://doi.org/10.1007/s11042-023-14493-z
Wu D, Yuan C (2022) Threshold image segmentation based on improved sparrow search algorithm. Multimed Tools Appl 81:33513–33546. https://doi.org/10.1007/s11042-022-13073-x
Chouhan SS, Kaul A, Singh UP (2018) Soft computing approaches for image segmentation: a survey. Multimed Tools Appl 77:28483–28537. https://doi.org/10.1007/s11042-018-6005-6
Bhargava A, Bansal A, Goyal V (2022) Machine learning-based automatic detection of novel coronavirus (COVID-19) disease. Multimed Tools Appl 81:13731–13750. https://doi.org/10.1007/s11042-022-12508-9
Sasmal B, Dhal KG (2023) A survey on the utilization of Superpixel image for clustering based image segmentation. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-14861-9
Lima T, Luz D, Oseas A, Veras R, Araujo F (2023) Automatic classification of pulmonary nodules in computed tomography images using pre-trained networks and bag of features. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-14900-5
Zhou S, Cheng Y, Tamura S (2014) Automated lung segmentation and smoothing techniques for inclusion of juxtapleural nodules and pulmonary vessels on chest CT images. Biomed Sign Process Control 13:62–70. https://doi.org/10.1016/j.bspc.2014.03.010
Shen S, Bui AA, Cong J, Hsu W (2015) An automated lung segmentation approach using bidirectional chain codes to improve nodule detection accuracy. Comput Biol Med 57:139–149. https://doi.org/10.1016/j.compbiomed.2014.12.008
Feng C, Wei H, Li M, Li X, Ding M (2018) An edge detection method for suspicious local regions in CT images with jaxtapleural nodules. MATEC Web Conf 232:02056. https://doi.org/10.1051/matecconf/201823202056
Moghaddam RM, Aghazadeh N (2023) Lung Parenchyma Segmentation from CT Images with a Fully Automatic Method. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-16040-2
Poonkodi S, Kanchana M (2023) Lung cancer segmentation from CT scan images using modified mayfly optimization and particle swarm optimization algorithm. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-15688-0
Dharmalingham V, Kumar D (2020) A model based segmentation approach for lung segmentation from chest computer tomography images. Multimed Tools Appl 79:10003–10028. https://doi.org/10.1007/s11042-019-07854-0
Yang J, Qiu K (2022) An improved segmentation algorithm of CT image based on U-net network and attention mechanism. Multimed Tools Appl 81:35983–36006. https://doi.org/10.1007/s11042-021-10841-z
Suji RJ, Godfrey WW, Dhar J (2023) Border to border distance based lung parenchyma segmentation including juxta-pleural nodules. Multimed Tools Appl 82:10421–10443. https://doi.org/10.1007/s11042-022-13660-y
Beula RJ, Wesley AB (2023) Accurate segmentation of lung nodule with low contrast boundaries by least weight navigation. Multimed Tools Appl 82:27635–27657. https://doi.org/10.1007/s11042-023-14620-w
Peng Y, Luan P, Tu H, Li X, Zhou P (2023) Pulmonary fissure segmentation in CT images based on ODoS filter and shape features. Multimed Tools Appl 82:34959–34980. https://doi.org/10.1007/s11042-023-14931-y
Chae SH, Moon HM, Chung Y, Shin J, Pan SB (2016) Automatic lung segmentation for large-scale medical image management. Multimed Tools Appl 75:15347–15363. https://doi.org/10.1007/s11042-014-2201-1
An FP, Liu Je (2021) Medical image segmentation algorithm based on multilayer boundary perception-self attention deep learning model. Multimed Tools Appl 80:15017–15039. https://doi.org/10.1007/s11042-021-10515-w
Afif M, Ayachi R, Said Y, Atri M (2023) Deep learning-based technique for lesions segmentation in CT scan images for COVID-19 prediction. Multimed Tools Appl 82:26885–26899. https://doi.org/10.1007/s11042-023-14941-w
Seelan LJ, Suresh LP (2019) A new framework for early detection and diagnosis of lung lesion using various classifiers. Int J Recent Technol Eng (IJRTE) 8(2):829–833. https://doi.org/10.35940/ijrte.B1166.0782S419
Author information
Authors and Affiliations
Contributions
Abdollah Khorshidi: Conceptualization, Project administration, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft and Revising.
Corresponding author
Ethics declarations
Conflicts of interest
The author has declared no conflicts of interest
Ethical approval
Not required
Ethics approval and consent to participate
Not applicable.
Consent for publication
The informed consent is Not Applicable.
Competing interests
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All processes including conception and design of the study, analysis and interpretation of the data, and also drafting and revising the manuscript have been contributed by Dr Abdollah Khorshidi who is expert in medial radiation subjects and engineering.
Highlights.
• Extended algorithm in region growing was projected to appoint the lung tumor edges.
• The extensive algorithm was carried out on different CT slices.
• A trade-off between accuracy and time was regarded.
• Starting the growth algorithm from multi-point created precise tumor edges.
• Dice coefficients were 0.79 and 0.88 for basilar and inclusive algorithms by p-value of 0.08.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khorshidi, A. Introducing extended algorithm for respiratory tumor segmentation. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18496-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18496-2