Abstract
Lung cancer constitutes the most severe cause of cancer-related mortality. Recent evidence supports that early detection by means of computed tomography (CT) scans significantly reduces mortality rates. Given the remarkable progress of Vision Transformers (ViTs) in the field of computer vision, we have delved into comparing the performance of ViTs versus Convolutional Neural Networks (CNNs) for the automatic identification of lung cancer based on a dataset of 212 medical images. Importantly, neither ViTs nor CNNs require lung nodule annotations to predict the occurrence of cancer. To address the dataset limitations, we have trained both ViTs and CNNs with three advanced techniques: transfer learning, self-supervised learning, and sharpness-aware minimizer. Remarkably, we have found that CNNs achieve highly accurate prediction of a patient’s cancer status, with an outstanding recall (93.4%) and area under the Receiver Operating Characteristic curve (AUC) of 98.1%, when trained with self-supervised learning. Our study demonstrates that both CNNs and ViTs exhibit substantial potential with the three strategies. However, CNNs are more effective than ViTs with the insufficient quantities of dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer stands as the foremost cause of cancer-related fatalities worldwide, with over 1.6 million newly diagnosed cases per year [1]. Computed tomography (CT) has been proposed as a valuable early screening tool for lung cancer. Typically, lung cancer appears as a nodule on a CT scan. Early differentiation between non-cancerous (benign) and cancerous (malignant) nodules holds paramount importance for clinical treatment. Computed-aided diagnosis (CAD) systems [2] are widely employed for nodule detection. However, nodule classification remains a challenging task for CAD systems.
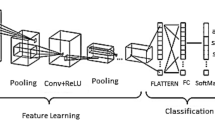
Current clinical classification algorithms require hundreds of images per patient, making it extremely challenging for a radiologist to perform precise and thorough analysis of all the images. To assist with disease diagnosis, radiomics [3] have gained popularity in extracting features from medical images, thanks to the advancements in computer systems [4, 5]. This identification scheme typically comprises four stages: preprocessing, segmentation, feature extraction, and classification. However, accurately segmenting lung lesion annotations for feature extraction in radiomics can be costly for doctors. Given the significant advancements in deep learning in recent years, we have investigated two deep learning approaches to identify lung cancer in this study. One of the key benefits of using deep learning methods is that the deep neural network considers context information from consecutive slices of a patient’s lungs, eliminating the need for manual segmentation of lung lesions, which is time-saving and convenient. As illustrated in Fig. 1, the three crucial stages in this study involve data preprocessing, feature extraction, and classification.
One of the approaches employed in this study involves the utilization of three-dimensional convolutional neural networks (3-D CNNs), which are considered the leading deep learning approach in automatic medical diagnosis [6,7,8,9]. The other approach utilizes vision transformers (ViTs) [10], which have been extensively utilized in natural language processing (NLP) and have emerged as a viable alternative to CNNs in computer vision, yielding comparable performance levels to those achieved by CNNs.
However, medical images are significantly different from natural images. For example, natural images are two-dimensional (2-D) RGB (Red, Green, Blue) images, while medical images are three-dimensional (3-D) grayscale images. Dosovitskiy et al. [10] trained and tested ViTs with 2-D natural images such as ImageNet [11]. Their results showed that ViTs could only outperform CNNs on very large-scale datasets. For instance, ViTs performed better after pre-training on JFT-300M, a 300 million-record proprietary dataset from Google. Thus, investigating the classification performance of ViTs on medical images is an urgent issue that needs to be addressed. Our research tackles the challenge of a relatively modest medical dataset consisting of 212 cases. This is a departure from the large-scale datasets traditionally used in ViTs experiments. The exploration of ViTs’ performance on a smaller dataset is a notable contribution, as it reflects real-world scenarios where medical data availability is often limited. To bridge the performance gap caused by limited data, we trained ViTs and CNNs using various methodologies to enhance their performance, as detailed in Section 2.2.
In essence, our research ventures into uncharted territory by evaluating the suitability of ViTs for medical image classification, acknowledging the data-specific challenges and offering various strategies to address them. It underscores the importance of adapting cutting-edge deep learning techniques for medical applications, ultimately contributing to the broader field of lung cancer identification.
2 Materials and methods
2.1 Model architecture
With the rapid development of deep learning in recent years, CNNs have been the controlling approach in the domain of computer vision, with applications ranging from classification [12,13,14,15,16], segmentation [17,18,19] and object detection [20,21,22,23]. In recent years, many deep neural network architectures have demonstrated outstanding performance in image classification tasks, such as VGG [24], GoogLeNet [25], ResNet [26], and EfficientNet [27]. When contemplating the implementation of a lung cancer identification system, EfficientNet3D emerges as a commendable option for processing 3-D medical images. This choice is supported by its comparable parameter count to ViT, along with its outstanding performance on the ImageNet dataset, establishing it as a state-of-the-art deep learning method.
2.1.1 EfficientNet
The inference accuracy and speed of a CNN-based model are greatly affected by CNN’s depth, width and resolution. Tan and Le [27] propose a new scaling method that uniformly scales all dimentions of depth/width/resolution using a simple yet highly effective compound coefficient. The scaling method equation can be described as:
where \(\alpha \), \(\beta \), and \(\gamma \) indicate how to assign these additional resources to the width, depth, and resolution of the network, and \(\phi \) is a user-specified coefficient that governs how many more resources are available for the model scaling.
Figure 2 shows the architecture of EfficientNet3D-b0 which we used in this work. EfficientNet3D consists of a baseline network model that is expanded by adding more layers, widening the network, and increasing the input image resolution. Compared to other popular architectures, this mixed-dimension scaling method allows EfficientNet3D to achieve higher accuracy with fewer parameters and lower computational costs. EfficientNet has achieved state-of-the-art results on multiple image classification tasks, including ImageNet, CIFAR-10, and CIFAR-100, and has been widely adopted in many computer vision applications. Overall, it is appropriate for the deployment of lung cancer identification system.
2.1.2 Vision transformers
ViTs have recently garnered significant interest in the realm of natural image processing, serving as a compelling alternative to CNNs. The transformer architecture, a fundamental deep neural network for processing sequential input data, has found widespread use in NLP. The transformer encoder framework can be characterized as a series of similar layers, as exemplified in Fig. 3. Each of these layers is comprised of two sub-layers: a multi-head self-attention mechanism and a simple multi-layer perceptron (MLP) of fully-connected feed-forward networks. In addition to these sub-layers, a residual connection [26] surrounds each one, culminating in layer normalization [28].
Attention function
As shown in Fig. 3, the query and key-value pairs can be translated into an output by the general attention function, where the query, key, value, and output are all vectors. The input of each layer in the encoder includes queries and keys with dimention \(d_k\), and values with dimention \(d_v\). It computes the dot products of the query with all keys, divided each by \(\sqrt{d_k}\), and computes the value’s dot products with the associated weights to get the output after applying a softmax function to get the weights on the value. In practice, a set of queries are packed together into a matrix Q as well as keys and values with matrices K and V respectively. The output of attention can be represented like:
where \(X_q, X_k, X_v\) are the inputs of transformer encoder, \(\odot \) denotes matrix dot product and \(W^Q, W^K, W^V\) are the parameter matrices for linear projection. If the Q, K and V are computed from the same input X, it is known as self-attention. Specifically, the multi-head self-attention can be described as:
where the linear projections are parameter matrices \(W^Q_i\in \mathbb {R}^{d_{model}\times d_k}\), \(W^K_i\in \mathbb {R}^{d_{model}\times d_k}\), \(W^V_i\in \mathbb {R}^{d_{model}\times d_v}\) and \(W^O_i\in \mathbb {R}^{hd_{v}\times d_{model}}\). For example, Deit-S [29] takes \(h = 6\) heads and for each head \(d_k = d_v = d_{model}/h = 64\).
The ViTs model has gained popularity in natural image processing alongside CNNs, owing to its ability to simultaneously examine data from multiple representation subspaces at various locations through multi-head attention. Furthermore, it provides an integrated saliency map that identifies areas of focus by the model, which is difficult to achieve with CNNs [30]. Additionally, the parallel attention heads structure allow for training on large-scale datasets and reduce training time.
An overview of the ViTs architecture is illustrated in Fig. 4. The first two layers are patch embedding and position embedding. In our methodology, we initially divided the CT images into patches of size 16X16X16, following the resampling process, which transformed the images into dimensions of 224X224X64. Subsequently, these patches were flattened into a single 1x4096-sized vector through patch embedding. The representation of the patch embedding is illustrated as follows:
where \(x^i\) is the \(i^{th}\) patch, \(\text {Flatten}(\cdot )\) denotes flattening the patch into 1X4096 size and \(\mathcal {F}(\cdot )\) denotes projecting the vector into 1X384 size.
Position embedding
In images, the spatial arrangement of pixels is crucial for understanding the content. Unlike text data, where the order of words is explicitly represented by their positions in the sequence, images inherently lack this positional information. Position embedding provides ViTs with a way to incorporate spatial relationships between pixels or patches in the image. It is typically added to the patch embedding and help the model understand the relative positions of different image elements. To learn the positional information between tokens, we add 1-D positional embedding for all the tokens which can be described as:
where \(q_{class}\) is class token, \(q^i\) is patch token and \(E_{pos}\) is a learnable matrice which is initialized with zero.
Class token
The class token is a unique token added to the patch embedding that serves as a global representation of the entire image. In NLP tasks, the transformer model typically uses a special token(e.g., [CLS]) to represent the entire sequence or document. Similarly, as shown in Fig. 4, the class token represents the overall content of the image. It provides a high-level context and is crucial for image classification tasks where the goal is to assign a single label to the entire image.
In image processing tasks, ViTs have several advantages compared to traditional CNNs:
-
1.
ViTs provide contextual features by utilizing self-attention mechanisms on input serialized data, which helps capture relationships between different positions in the input, leading to more accurate image processing.
-
2.
ViTs’ parallelized network architecture allows them to be trained on larger datasets and significantly reduces training time. This enables ViTs to efficiently handle large-scale image datasets.
-
3.
ViTs come with built-in attention maps, which provide insights into the regions of interest that the model focuses on when making decisions. This enhances interpretability and helps understand how the model operates.
Applying ViTs to computer vision has potential advantages as it allows the use of the same deep neural network model for both computer vision and natural language processing tasks, facilitating developments in the multimodal field. Therefore, exploring the classification performance of ViTs on medical images is of great significance, as it is expected to offer improved performance and interpretability, thereby enhancing the analysis and diagnosis of medical images.
2.2 Training strategies
ViTs have been widely used in the field of natural image processing, but their utility in the field of medical images is limited since the medical datasets typically have insufficient quantities and frequently come with less trustworthy labels. Therefore, in order to enhance the performance of CNNs and ViTs, we additionally investigate three training methodologies include transfer learning, self-supervised learning and sharpness-aware minimizer.
The first approach is transfer learning, which entails utilizing pre-existing weights that have undergone training on ImageNet and fine-tuning for optimal performance on the own dataset. According to Raghu et al. [31], transfer learning has been demonstrated to increase the overall efficacy of the model, as well as to significantly reduce the duration of the training process. However, the scarcity of ample datasets for 3-D medical images imposes constraints on the application of transfer learning for 3-D CNNs, because ImageNet is of 2-D natural images. As an alternative, we can adopt the use of ViTs, where only the patch embedding and position embedding are initialized from scratch, while the remaining layers are initialized with pre-trained weights from ImageNet.
The imaging shows of each lung cancer categories. (a) AIS: a ground-glass nodule in the lateral segment of the right middle lung lobe. (b) MIA: a ground-glass density foci in the posterior segment of the left upper lung lobe. (c) IAC: a ground-glass nodule in the apical segment of the right upper lung lobe. (d) Hamartoma: a round high-density nodule in the dorsal segment of the right lower lung lobe. (e) Inflammatory pseudotumor: a nodule in the dorsal segment of the left lower lung lobe. (f) Atypical hyperplasia: a ground-glass density nodule in the anterior medial basal segment of the left lower lung lobe. (g) Granuloma: an anterior segment nodule in the apical segment of the right upper lung lobe. (h) Benign tumor: a lobular nodule in the basal segment of the right lower lung lobe. (i) Pneumonia: a soft tissue nodule in the lateral segment of the right middle lung lobe
Recent advances in self-supervised learning (SSL) have improved the deep neural network’s ability to express unlabeled data. For instance, BYOL [32] utilizes two neural networks, namely the online and target networks, which interact and learn from each other, and achieves a state-of-the-art performance on ImageNet using a standard ResNet-50 [33]. Hua et al. [34] propose a progressive hierarchical feature alignment method to solve the imbalance problems of data and classes. Michieli et al. [35] propose an Unsupervised Domain Adaptation strategy to learn on labeled synthetic data and unlabeled real data. Their results prove the effectiveness of the proposed strategy in adapting a segmentation network trained on synthetic datasets to real world datasets. The difference between ssl and them is that we do unsupervised training on a real world dataset and then use the learned knowledge in supervised learning tasks. DINO [36] uses self-distillation to encourage a student and teacher network to produce similar representations given inputs of different augmentations, and achieves the highest performance on ImageNet and other standard benchmarks with ViTs. Therefore, we also employ SSL to train all models and assess whether it benefits the identification of lung cancer.
Furthermore, it should be noted that the training of ViTs is sensitive to initialization and hyperparameter selection, as pointed out by Touvron et al. [29], which may result in low training error but high generalization error during evaluation [37, 38]. To address this issue, Chen et al. [39] propose the use of the Sharpness-Aware Minimizer (SAM) optimization technique to achieve smoother loss landscapes for each network component and higher weight norms in ViTs. Additionally, ViTs trained from scratch on ImageNet, with limited pre-training and data augmentation, have demonstrated superior performance compared to similarly sized ResNets, aligning with the characteristics of 3-D medical image datasets. Thus, we also incorporate the SAM optimization method to improve the accuracy of lung cancer identification.
2.3 Dataset
As outlined in Table 1, our dataset was meticulously collected from the Shandong Provincial Hospital, affiliated with Shandong First Medical University, spanning the period from September 2018 to December 2020. This comprehensive dataset comprises a total of 212 cases, consisting of 130 malignant and 82 benign tumor cases. Figure 5 provides a visual representation of the lesions pertaining to each lung cancer category. Within this figure, the red circle is employed to denote the largest lesion in the dataset, based on its diameter.
Each entry in our dataset corresponds to a CT scan with dimensions of \(512\times 512 \times z\), where z represents the number of slices acquired for each scan. It’s important to note that all the cases included in our dataset exhibited imaging findings indicative of lung cancer, warranting the necessity for surgical resection. The lesions observed in these cases exhibited a range in terms of their longest diameter, spanning from 0.6 cm to 3 cm. Moreover, it is imperative to highlight that each of these lesions underwent surgical removal and was subsequently confirmed through pathological examination.
With regard to the classification of lung tumors in our dataset, they can be categorized as follows:
-
Malignant. If the tumor is malignant, it is an uncontrollable level of cancer tumor and is no cure in this category.
-
Benign. If the tumor is benign, it is starting level of cancer tumor and is easy to cure in this category.
3 Experiments
3.1 Preprocessing
The preprocessing of lung CT scans is undertaken with the aim of improving the quality of the scans, thus achieving superior outcomes in the diagnosis of lung cancer. As depicted in Fig. 6, the Hounsfield scale (HU) of the CT scans was adjusted to fall between -1000 and 1000, after which normalization was carried out to maintain image values within the range of 0 to 255. Subsequently, all CT scans were resized to size of 224X224X64, following which zero centering was applied to each slice of the scans. The zero centering is described as (6):
where \(\mu \) is the mean intensity of one slice and \(\sigma \) is the standard deviation intensity of one slice. A 7 : 3 random split was made for the training and validation. We also used RandomAnisotropy, random VerticalFlip, random HorizontalFlip, RandomBlur and RandomAffine as the medical data augmentations [40] before training the models.
3.2 Training settings
We have opted DeiT-S [29] as the transformer-based model which is identical to the ViTs but has been implemented in Pytorch [41]. In addition, we have chosen EfficientNet3D-b0 [27] as the 3-D CNN-based model, as Deit-S and EfficientNet3D-b0 have similar efficiency and parameter counts. The hyperparameters for training are presented in Table 2. Previous studies have demonstrated the advantages of using “LinearWarmup” [42] to train transformers. Hence, we have utilized “LinearWarmup” of 10 epochs with an initial learning rate of 1X\(10^{-8}\) and “CosineAnnealingLR” [43] with a final learning rate of 1X\(10^{-6}\) as the learning rate schedulers. Each experiment was conducted on a single host equipped with two TITAN RTX GPUs (24GB memory).
3.3 Evaluation metrics
In order to evaluate the different models against each other, we choose accuracy, precision, recall, F1 score and AUC as the metrics for the lung cancer identification. Accuracy is the percentage of correct predictions out of all cases evaluated, which is described as:
where TP, TN, FP and FN are true positive, true negative, false positive and false negative of the confusion matrix [44]. Precision is described as
and it refers to the proportion of truely positive among the number of positive predictions. Recall is described as
and it describes the proportion of the correctly classified among the actual positive cases. The F1 score, which ranges from 0 to 1, represents the harmonic mean of recall and precision and is described as follows:
The area under the receiver operating characteristic curve, or AUC [45], demonstrates how well the probabilities from the positive classes are separated from the negative classes. The ROC curve is plotted by false positive rate (FPR) and true positive rate (TPR) with various threshold values, where FPR is simply the percentage of erroneous detections that the algorithm is catching and can be calculated as (1 - specificity). It can be described as:
TPR is just the percentage of trues that the algorithm is catching and is also known as sensitivity or recall, which can be described as:
4 Results
In Table 3, we compare Deit-S against EfficientNet3D using different training strategies. Initially, the AUC (95.2%) of EfficientNet3D greatly exceeded that of Deit-S (89.2%) when trained with randomly initialized weights (Kaiming initialization [46]). However, in medical imaging recognition tasks, random initialization is no longer the standard practice. Instead, the standard procedure is transfer learning. As shown in Table 3, transfer learning increased the AUC of Deit-S from 89.2% to 95.3%. This suggests that Deit-S benefited significantly from transfer learning for lung cancer identification, and achieved similar performance to EfficientNet3D.
We also compared Deit-S against EfficientNet3D with SSL and SAM in Table 3. Results showed that pre-training EfficientNet3D with SSL increased its AUC from 95.2% to 98.1%, and Deit-S from 89.2% to 95.7%. Additionally, SAM improved the AUC of EfficientNet3D from 95.2% to 97.4% and Deit-S from 89.2% to 96.5%. These findings suggest that both SSL and SAM can successfully enhance the capabilities of ViTs and CNNs for lung cancer identification, with ViTs exhibiting more improvement than CNNs under these settings.
Moreover, Fig. 7 revealed that EfficientNet3D achieved the highest score of AUC (98.1%) when pre-trained with SSL, and the highest recall score (94.7%) when trained with SAM. However, ViTs could not achieve the same level of performance as CNNs, even when trained with SSL or SAM.
5 Discussion
The results presented in Table 3 highlight the significant impact of different training strategies on the performance of Deit-S and EfficientNet3D in the task of lung cancer identification. Initially, when both models were trained with randomly initialized weights, EfficientNet3D outperformed Deit-S. However, when Deit-S was fine-tuned using transfer learning with pre-trained weights from ImageNet, its AUC improved substantially, reaching 95.3%. This suggests that Deit-S can benefit significantly from transfer learning in the context of lung cancer identification and achieve performance comparable to EfficientNet3D.
The comparison between Deit-S and EfficientNet3D with SSL and SAM further illuminates the impact of advanced training techniques. Pre-training both models with SSL resulted in substantial improvements in AUC. Additionally, the incorporation of SAM led to enhanced AUC scores for both models, with EfficientNet3D achieving 97.4% and Deit-S reaching 96.5%. These findings indicate that SSL and SAM can effectively enhance the capabilities of both ViTs and CNNs for lung cancer identification. Notably, ViTs exhibited a more significant improvement in performance compared to CNNs under these settings.
Figure 7 provides additional insights into the models’ performance, particularly in terms of confusion matrix. EfficientNet3D pre-trained with SSL achieved the highest AUC of 98.1%, demonstrating its potential when leveraged with SSL techniques. On the other hand, when trained with SAM, EfficientNet3D exhibited the highest recall score of 94.7%, indicating its superior ability to detect lung cancer cases. However, it is essential to highlight that ViTs, including Deit-S, did not achieve the same level of performance as CNNs, even when trained with SSL or SAM. This discrepancy in performance could be attributed to the limited availability of training data. It aligns with the theory that ViTs, lacking inherent “inductive bias”, may struggle to generalize effectively when trained with a restricted dataset.
In summary, the results and discussions presented herein underscore the importance of training strategies in the performance of deep learning models for lung cancer identification in medical imaging. Transfer learning, SSL, and SAM have been demonstrated to be valuable tools for enhancing the capabilities of both ViTs and CNNs, though the training of ViTs is sensitive to initialization and hyperparameter selection. While ViTs show promise, their performance may be limited by data availability, highlighting the need for further research into mitigating this limitation and harnessing the full potential of ViTs in medical image analysis.
6 Interpretability of transformer
The Transformer-based model presents a unique advantage in its inherent interpretability, a feature of paramount significance for comprehending its inner workings and the rationale behind its predictions. This interpretability is chiefly facilitated through the generation of saliency maps of attention, which effectively highlight the pivotal regions within the input image that contribute significantly to the model’s decision-making process. These saliency maps are created by computing the self-attention weight scores associated with a transformer encoder layer, where these weight scores essentially quantify the degree of emphasis placed on each individual image patch in relation to its counterparts.
To delve into the specifics of the saliency map generation process, we specifically extracted the attention weights from the final self-attention block of the transformer encoder, as depicted in Fig. 4. These attention weights serve as a testament to the model’s attentional focus and are instrumental in crafting the saliency maps. By computing the weighted sum of the feature maps corresponding to each image patch, we effectively obtain a two-dimensional saliency map with dimensions mirroring those of the input image patches.
As illustrated in Fig. 8, the resultant saliency maps generated for ViTs when trained with SSL reveal some intriguing insights. Notably, each attention head exhibits subtle variations in the patches it prioritizes during the decision-making process. However, it is evident that, in the broader context, ViTs tend to concentrate their attention predominantly on the lung region when making predictions. This phenomenon aligns seamlessly with the distribution of tumor regions, which tend to be densely clustered within the lungs. Such nuanced attention maps, showcasing distinct areas of focus across different heads and yet an overarching emphasis on medically relevant regions, present a remarkable aspect of interpretability unique to ViTs. This level of interpretability is challenging to attain when employing CNNs-based models.
This facet of interpretability assumes paramount importance in the context of employing artificial intelligence for medical diagnosis. It not only assists in demystifying the model’s decision-making process but also empowers researchers and medical practitioners with invaluable insights into the reasoning and justifications underlying the AI model’s predictions. This enhanced transparency aids in validating the model’s accuracy and facilitates fine-tuning and optimization for more robust and reliable medical applications.
7 Conclusions
In the present study, we undertook the task of developing an automated system for early lung cancer identification, employing two prominent deep learning approaches: Deit-S and EfficientNet3D. Moreover, we delved into the utilization of critical training strategies, including transfer learning, self-supervised learning, and sharpness-aware minimizer, to enhance the performance of both ViTs and CNNs. Our research outcomes underscore the efficacy of these techniques in elevating the predictive power of these models, with CNNs achieving the highest AUC of 98.1% when trained with SSL. This achievement highlights the potential of CNNs in identifying lung cancer with exceptional accuracy.
Nonetheless, it is crucial to acknowledge that our study also reveals certain limitations in the current state of ViTs for medical imaging tasks. Despite their promise, ViTs did not match the performance levels of CNNs in our experiments. This discrepancy emphasizes the need for further research and development in leveraging ViTs effectively for medical image analysis. It suggests that ViTs might require more substantial data resources or novel architectural adaptations to fully exploit their capabilities in this domain. Looking ahead, future research efforts should concentrate on training ViTs and CNNs on larger-scale lung cancer datasets and other CNN models. The expectation is that, as the dataset size increases, ViTs will progressively bridge the performance gap with CNNs and potentially surpass them in accuracy and robustness. This evolution promises to bring significant benefits to medical practitioners by enabling more precise and early diagnosis of lung cancer, ultimately saving lives through earlier interventions.
In conclusion, our study has shed light on the immense potential of deep neural networks, particularly CNNs, for the task of lung cancer identification based on medical CT images. We have demonstrated the impact of advanced training strategies and highlighted the ongoing challenges faced by ViTs in this context. The journey towards harnessing the full potential of ViTs in medical imaging is an exciting frontier that holds the promise of even more accurate and accessible healthcare solutions in the future.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Tsao AS, Scagliotti GV, Bunn PA Jr, Carbone DP, Warren GW, Bai C, De Koning HJ, Yousaf-Khan AU, McWilliams A, Tsao MS (2016) Scientific advances in lung cancer 2015. J Thor Oncol 11(5):613–638
Fraioli F, Serra G, Passariello R (2010) CAD (computed-aided detection) and CADX (computer aided diagnosis) systems in identifying and characterising lung nodules on chest CT: overview of research, developments and new prospects. La Radiol Med 115(3):385–402
Kukreja V, Sakshi (2022) Machine learning models for mathematical symbol recognition: a stem to stern literature analysis. Multimedia Tools Appl 81(20):28651–28687
Vijaya G, Suhasini A, Priya R (2014) Automatic detection of lung cancer in CT images. IJRET: Int J Res Eng Technol 3(7):182–186
Sakshi, Kukreja V (2023) A dive in white and grey shades of ml and non-ml literature: a multivocal analysis of mathematical expressions. Artif Intell Rev 56(7):7047–7135
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imag 35(5):1299–1312
Aurna NF, Yousuf MA, Taher KA, Azad A, Moni MA (2022) A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput Biol Med 146:105539
Rostami B, Anisuzzaman D, Wang C, Gopalakrishnan S, Niezgoda J, Yu Z (2021) Multiclass wound image classification using an ensemble deep CNN-based classifier. Comput Biol Med 134:104536
Deepak S, Ameer P (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med 111:103345
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. arXiv:2010.11929
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. Paper presented at the 2009 IEEE conference on computer vision and pattern recognition, pp 248–255
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inform Process Syst 25
Hershey S, Chaudhuri S, Ellis DP, Gemmeke JF, Jansen A, Moore RC, Plakal M, Platt D, Saurous RA, Seybold B (2017) CNN architectures for large-scale audio classification. Paper presented at the 2017 IEEE international conference on acoustics, speech and signal processing (icassp), pp 131–135
Bardou D, Zhang K, Ahmad SM (2018) Lung sounds classification using convolutional neural networks. Artif Intell Med 88:58–69
Kukreja V, Lodhi S et al (2023) Impact of varying strokes on recognition rate: a case study on handwritten mathematical expressions. Int J Comput Digit Sys
Kukreja V (2021) A retrospective study on handwritten mathematical symbols and expressions: classification and recognition. Eng Appl Artif Intell 103:104292
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Paper presented at the international conference on medical image computing and computer-assisted intervention, pp 234–241
Sakshi Kukreja V (2023) Image segmentation techniques: statistical, comprehensive, semi-automated analysis and an application perspective analysis of mathematical expressions. Archiv Computat Methods Eng 30(1):457–495
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. Paper presented at the international conference on medical image computing and computer-assisted intervention, pp 424–432
Girshick R (2015) Fast R-CNN. In: Proceedings of the IEEE international conference on computer vision, pp 1440–1448
Ren S, He K, Girshick R, Sun J (2015) Faster R-CNN: towards real-time object detection with region proposal networks. Adv Neural Inform Process Syst 28
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 779–788
Rosati R, Romeo L, Silvestri S, Marcheggiani F, Tiano L, Frontoni E (2020) Faster R-CNN approach for detection and quantification of DNA damage in COMET assay images. Comput Biol Med 123:103912
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning, pp 6105–6114
Ba JL, Kiros JR, Hinton GE (2016) Layer normalization. arXiv:1607.06450
Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H (2021) Training data-efficient image transformers & distillation through attention. In: International conference on machine learning, pp 10347–10357
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A.N, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inform Process Syst 30
Raghu M, Zhang C, Kleinberg J, Bengio S (2019) Transfusion: understanding transfer learning for medical imaging. arXiv:1902.07208
Grill J-B, Strub F, Altché F, Tallec C, Richemond P, Buchatskaya E, Doersch C, Avila Pires B, Guo Z, Gheshlaghi Azar M (2020) Bootstrap your own latent-a new approach to self-supervised learning. Adv Neural Inform Process Syst 33:21271–21284
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision, pp 630–645
Hua Y, Yi D (2021) Synthetic to realistic imbalanced domain adaption for urban scene perception. IEEE Trans Ind Inform 18(5):3248–3255
Michieli U, Biasetton M, Agresti G, Zanuttigh P (2020) Adversarial learning and self-teaching techniques for domain adaptation in semantic segmentation. IEEE Trans Intell Veh 5(3):508–518
Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, Joulin A (2021) Emerging properties in self-supervised vision transformers. arXiv:2104.14294
Hendrycks D, Dietterich T (2019) Benchmarking neural network robustness to common corruptions and perturbations. arXiv:1903.12261
Hendrycks D, Basart S, Mu N, Kadavath S, Wang F, Dorundo E, Desai R, Zhu T, Parajuli S, Guo M (2021) The many faces of robustness: a critical analysis of out-of-distribution generalization. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 8340–8349
Chen X, Hsieh C-J, Gong B (2021) When vision transformers outperform resnets without pretraining or strong data augmentations. arXiv:2106.01548
Pérez-García F, Sparks R, Ourselin S (2021) Torchio: a python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Comput Methods Programs Biomed 106236. https://doi.org/10.1016/j.cmpb.2021.106236
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L et al (2019) Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inform Process Syst 32
Liu L, Jiang H, He P, Chen W, Liu X, Gao J, Han J (2019) On the variance of the adaptive learning rate and beyond. arXiv:1908.03265
Loshchilov I, Hutter F (2016) SGDR: Stochastic gradient descent with warm restarts. arXiv:1608.03983
Powers DM (2020) Evaluation: from precision, recall and f-measure to roc, informedness, markedness and correlation. arXiv:2010.16061
Fawcett T (2006) An introduction to roc analysis. Pattern Recogn Lett 27(8):861–874
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision, pp 1026–1034
Acknowledgements
This study was supported by the Natural Science Foundation of China under Grant U1806202, Grant 61533011 and the Natural Science Foundation of Shandong Province of China ZR2019BF035, ZR2020ZD25, ZR2021QF042, 2022CXGC10501.
Funding
Natural Science Foundation of Shandong Province of China (ZR2021MF057).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that we have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gai, L., Xing, M., Chen, W. et al. Comparing CNN-based and transformer-based models for identifying lung cancer: which is more effective?. Multimed Tools Appl 83, 59253–59269 (2024). https://doi.org/10.1007/s11042-023-17644-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17644-4