Abstract
The early diagnosis of cancer is crucial to provide prompt and adequate management of the diseases. Imaging tests, in particular magnetic resonance imaging (MRI), are the first preferred method for diagnosis. However, these tests have some limitations which can cause a delay in detection and diagnosis. The use of computer-aided intelligent systems can assist physicians in diagnosis. In this study, we established a Convolutional Neural Network (CNN)-based brain tumor diagnosis system using EfficientNetv2s architecture, which was improved with the Ranger optimization and extensive pre-processing. We also compared the proposed model with state-of-the-art deep learning architectures such as ResNet18, ResNet200d, and InceptionV4 in discriminating brain tumors based on their spatial features. We achieved the best micro-average results with 99.85% test accuracy, 99.89% Area under the Curve (AUC), 98.16% precision, 98.17% recall, and 98.21% f1-score. Furthermore, the experimental results of the improved model were compared to various CNN-based architectures using key performance metrics and were shown to have a strong impact on tumor categorization. The proposed system has been experimentally evaluated with different optimizers and compared with recent CNN architectures, on both augmented and original data. The results demonstrated a convincing performance in tumor detection and diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Brain tumors are one of the most common causes of human death. Thus, an early and accurate diagnosis is critical for an effective treatment process. In clinical neuroradiology, pre-treatment diagnosis of brain tumors using Magnetic Resonance Imaging (MRI) is challenging. This is because in contrast t-1 weighted MRI scans, their appearance is very similar to hyperintense brain lesions such as lipoma, dermoid cysts, thrombosis, etc. [65]. Additionally, the human eye cortex has a limited capacity to distinguish between different gray levels present in both MRI and computerized tomography.
As brain cells renew themselves, the abnormal cells that occur in the replication phase grow and become a mass, forming brain tumors. There are two types of brain tumors with benign/primary (e.g. pituitary or meningioma) and malignant/secondary (e.g. glioma). While benign brain tumors do not spread, malignant tumors spread throughout the body using different organs, such as breasts and lungs to make brain metastases [48]. One type of malignant tumors, gliomas not only invade the surrounding tissues but have the ability to metastasize to distant tissues. As a result, they require a quick and accurate diagnosis, as they have a faster growth rate, a tendency to invade surrounding tissues, and the ability to metastasize to different tissues. Therefore, the development of more effective therapeutics in both diagnosis and treatment is crucial. Additionally, there are some drawbacks to the ability of conventional MRI to discriminate between primary and metastases tumors and central nervous system masses, because their radiological features appear to be similar. Artificial Intelligence (AI)-based research on existing data is required to help guide decisions, comprehensive datasets from various users, and retrospective analysis of data to shed light on exploring new avenues in both diagnostic and therapeutic processes [12].
With the development of computer vision technology, AI technology produces smart solutions in many fields such as industry [43, 70], medicine (e.g. cancer detection) [20], early diagnosis and treatment of non-symptomatic liver disease [41], predicting fatal malaria [55], thoracic surgery [21], face mask detection for COVID-19 prevention [5, 52, 54], nanotechnology [13], robotics [19, 72], agriculture [53, 56, 74]. Convolutional Neural Network (CNN) is one of the most commonly used neural networks in Deep Learning (DL) with its strong self-learning, adaptability, and generalization ability.
This article proposes a new CNN-based framework to detect tumors and categorize brain tumor types at the pre-diagnosis stage. In the proposed framework, the MR images are first cropped up to the skull, then the histogram equalization and denoising filter are performed. Then, the data augmentation technique is used to ensure stable learning. Finally, the EfficientNetv2 + Ranger pre-trained CNN model was performed using the fine-tuned hyperparameters for brain tumor detection and pre-diagnosis.
The main contributions of this study are:
-
1
To introduce a new CNN-based classification system with EfficientNetv2 + Ranger architecture for malign tumors (glioma, meningioma, and pituitary).
-
2
To show that data pre-processing is crucial for the accurate diagnosis of tumors.
-
3
To provide better accuracy and stable learning procedure of the automated diagnosis of tumors compared to the performance of the other CNN architectures in multi-class scenarios using Ranger optimizer on different datasets.
-
4
To explore the behaviors of recent optimization algorithms in CNN networks for MRI images.
-
5
To propose an alternative method for the rapid diagnosis of brain tumors using computer-assisted radiological examination in addition to neurological examination, and a future-oriented guide intended to encourage other scientists to conduct advanced studies in this area.
In the general structure of this work, Section 2 reviews related works. Then, a detailed description of the basic algorithm, with some considerations on the pre-processing tasks, is presented in Section 3. In Section 4, the experiments are evaluated using both the perceptual and classical quality criteria and the effectiveness of our methodology. Section 5 discusses the detailed results using existing tumor classification methods and Section 6 introduces the conclusions and future work, respectively.
2 Related works
Computer-assisted smart healthcare systems have been rapidly developing in order to provide more coordinated and quality service to patients needing after a disease diagnosis. After the emergence of MRI, it became possible to analyze both the anatomical status and the biochemical structure of brain morphology. Additionally, CNN-based methods have made tremendous progress in analyzing the growing quantity and diversity of distinct tumor types. [27] compared different CNN architectures where these two CNN architectures used to solve these problems related to the instability of tumor labels using a two-step training procedure. Unlike traditional CNN models, the two-step training procedure was constructed using the average of the outputs from both local details and global texture features. [71] solved the medical image classification problem by using features from segmentation networks, which allowed for the learning procedure to be executed more easily and robustly in real classification problems involving complex structures. Their study compared the pre-trained ImageNet classifiers and the scratch-trained classifiers and they demonstrated that the ImageNet and pre-trained VGGNet [64] neural network yielded more successful results. Similarly, [38] developed a new abnormal brain disease categorization using several classes i.e., sarcoma, meningioma, gliomas, and metastases through the transfer learning method and VGGNet neural network. In this approach, the last few layers of VGGNet were updated to embed new image categories by including a pre-trained model in the learning procedure. Although this model has a very long training time, it has slightly better accuracy than existing studies focused on standard performance metrics. [40] established an algorithm that classifies brain images either as tumor or non-tumor using ensemble learning techniques based on pre-processing, segmentation, and feature extraction. As an alternative, [6] improved an Extreme Learning Machine (ELM)-based method to use when evaluating model performance by synthesizing cropped, uncropped, and segmented lesions of different dimensions of t1-weighted scans [16]. In [51], all traditional machine learning algorithms and the CNN methods enhanced for brain tumor detection were broadly compared including feature extraction and classification. Unlike the other studies, [58] described a brain tumor diagnosis algorithm, which included image segmentation based on the Unet architecture using advanced CNN architectures. In comparison to the previous approaches, this new strategy was built on VGG16 network architecture as the backbone of the Unet architecture. In particular, since clinical applications may include patients with different pathological tumor findings, the results should be accurate and obtained within a narrow time period. Advances in various experimental imaging techniques, including the early detection of brain abnormalities, are helping to increase the popularity of MRI as a complementary modality for treatment in the patients' clinical preparations for treatment. Another approach is to use an evolutionary algorithm to adapt the reinforcement learning to classify and detect brain tumors [60]. This involves two phases of pre-processing: freezing and fine-tuning. The significant features are then extracted from the MRI slices. However, this approach is not applicable when inaccurate segmentation has been done as it depends directly on the segmentation performance. As an alternative to the previously mentioned research, [4] found that a CNN technique based on the RELU-derived hard swish activation function could better extract CNN edge and texture features in order to detect cancerous tissue. Many powerful CNN models have recently accomplished perfect progress in computer-assisted clinical programs, including segmentation [9, 34], diagnosis, and the classification [8, 59, 69] of medical images using radiological data.
In terms of the weights, bias, and other learning parameters, ResNet50 [28], DenseNet201 [33], MobileNetv2 [31], InceptionV3 [66], and NASNet [75] artificial neural network layers are capable of high-level brain tumor identification and learning a large number of key features. One of the major drawbacks of these proposed systems is the small number of training data used to categorize brain tumors. Additionally, some CNN architectures such as VGGNet, KE-CNN, and ResNet50 may perform insufficiently in both tumor detection and classification tasks when determining the subtle alterations of the brain morphology on the MRI. Some of these differences include total brain volume, corpus callosum, increases in the total white matter volume, etc. These architectures can easily learn the underlying data patterns, which can lead to overfitting, poor generalization, and difficulty in interpreting the results. In this study, in order to address these drawbacks, we pre-processed the whole dataset before feeding them to the neural network. This step worked to prevent overfitting and under-fitting, which could affect the performance. Meanwhile, [67] proposed a novel EfficientNetv2 CNN family with lower training time and a better efficiency than previous models. This study shows that careful balancing not only in hyper-parameters but also network depth, width, and resolution in a CNN model leads to better performance. However, we provided a better balance of weight and bias by minimizing the error using a different optimizer, Ranger, in forward and backward propagation, to better extract the features of the tumor region. Based on the related studies, 4 state-of-the-art CNN models EfficientNetv1, ResNet18, ResNet200d, and InceptionV4 were included in this study to ensure diagnosis and multiple classifications of similar brain tumor diseases in addition to the EfficientNetv2 architecture.
3 Materials and methods
Recent years have witnessed fast development in CNN technologies. CNNs have become popular due to their high sensitivity and their ability to be used in a wide range of applications and research areas such as signal processing [1, 35, 47], pattern recognition [36], authentication systems [2, 29, 50]. CNNs are an advanced concept of artificial neural networks and have gained increasing attention due to both their learning stability and capability of processing images with varying quality for computer vision problems in many different fields. In our approach, we used a CNN-based system to diagnose 3 different malignant brain tumors using the EfficientNetv2 model powered by the state-of-the-art optimizer Ranger and fine-tuned pre-processing. At the time of publication, to the best of our knowledge, this idea has not been considered in the literature. Based on the achievement of CNNs in solving various complex tasks, this improved framework aims to achieve optimal multi-class brain tumor diagnosis by clarifying the missing details in MRI images against the restrictions such as noise, blur, and brightness. Figure 1 shows the flow diagram of the proposed system. The system includes the following stages: 1. Pre-processing, 2. Data generation, 3. CNN framework or deep feature extraction for tumor detection and 4. Diagnosis. In the first step, the image datasets were cropped and filtered against image impairments. The next process was data augmentation and the whole dataset was balanced using different transformation and noise invariance methods using different parameters. Just after splitting the dataset into training, validation, and test sets, the input was sent to the trained model on MR images for the detection of cancer.
3.1 Datasets
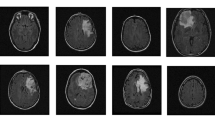
In this work, to ensure a less dataset-specific and more generalizable approach, two publicly-available datasets BR35H::Brain Tumor Detection 2020 (BR35H) [26] and t1-weighted contrast-enhanced MRI dataset [16] were chosen. The first dataset consisted of a total of 2768 different resolution MR images, of which 1458 images showed different tumor types and the remaining 1310 images showed healthy brains. The second dataset included CE-MR slices of 3064 t1-weighted meningiomas, gliomas, and pituitary tumors, and each MR slice had a size of 512 × 512 resolution (pixel dimensions 0.49 × 0.49 mm2). The used images in this study are illustrated in Fig. 2 and the tumor borders are highlighted in red.
3.2 The EfficientNetv2 architecture
In recent scientific studies, the EfficientNetv2 model has been preferred as a powerful tool (faster training speed and better parameter efficiency) in smart healthcare systems, as it is capable of handling medical image analysis successfully. It has been used for automatic tuberculosis diagnosis in chest X-ray images [3], breast cancer classification [62], and COVID-19 detection using X-ray and CT images [32].
Although the EfficientNetv2 architecture was developed the same way as EfficientNetv1, EfficientNetv2 is generally superior to EffiecientNetv1 in terms of parameters and floating-point operations per second (FLOP) efficiency. The FLOPs indicate the complexity of the model by measuring the number of transactions of a frozen CNN network. Due to EfficientNetv1 being trained with a large image size, it consumes a significant amount of memory. Since the total memory size in the Graphics Processing Unit (GPU) is limited, it is necessary to execute CNN models with a smaller batch size, which slows the training speed down considerably. One of the superior features of EfficientNet compared to the other models is its depth-wise convolutions [63].
In the EfficientNetv2 architecture, the original Inverted Residual Block (MBConv) in EfficientNet-B4 was replaced with Fused-MBConv in the first layers. In this architecture, a non-uniform scaling strategy was used to gradually add more layers to subsequent stages. Through progressive training, it initially provides low regularization by using small images during training which is then followed by high regularization. Through this process it achieves a positive impact on the width or depth expansion of the network and extracts the discriminative features. Thus, it achieves better model performance with fewer parameter sizes. This model, with shallow and faster CNN for image recognition, was developed by optimizing with a non-uniform expansion procedure. Then step-by-step extra layers were added to the next phase for sensitive training. In addition, the scaling rule was modified and the maximum image size was limited to a smaller value. The training speed increased while the parameter compatibility was maintained.
3.3 Comparison of Ranger with current optimizers
Optimization Function (OF) is a critical component in CNN networks and the performance of the optimization algorithm directly impacts the training efficiency of a model. However, the compatibility of different optimization algorithms with different CNN models can be achieved through the optimization of the cost function by adapting the neural network’s attributes such as learning rate, weights, and bias. To obtain optimal results from the network, we used the cutting-edge optimizer Ranger [68], instead of the default Adam optimizer [39], for the EfficientNetv2.
The Ranger optimizer combines two mechanisms: the Rectified Adam (RAdam) [44] and the stochastic optimizer LookAhead [49]. The RAdam was built by first determining an automatic warm-up mechanism using a rectifier function based on the actual variance encountered and then stabilizing learning at the beginning of training.
Based on the variance, the RAdam rectifies both the variance and generalization issues in the Adam stochastic optimizer's adaptive momentum. The main reason for these issues is that in the early steps of model training, the adaptive learning rate has an undesired large variance due to the limited amount of training dataset being used. Fundamentally, there is a close relationship between stability and variance.
By keeping an exponential moving average of the weights updated every 5 steps and replaced with the existing weights, LookAhead stabilizes learning and convergence during the rest of the training. LookAhead improves the network stability and reduces the variance of its inner optimizer with negligible computation and memory cost. Thus, it ensures a robust and stable breakthrough during the training period, as shown in Fig. 3. In CNN networks, the correct extraction of the most representative features depends heavily on the stability of the optimizer, which reducing the variance in the process. Therefore, it is possible to obtain higher accuracy rates by combining Ranger and EfficientNetv2.
4 Experiments
In this section, the experimental studies and a detailed evaluation of the improved system are discussed. Figure 4 illustrates the CNN-based tumor diagnosis system for brain tumors. During the study multiple CNN architectures and modern optimizers were trained and tested in order to identify the best-suited network. All networks were pre-trained using the ImageNet database. To construct the optimal model, we employed a fivefold cross-validation (CV) method, where each fold was divided into the subset (test: 10%, train: 80%, and validation: 10%) for every 10 iterations. We initially assigned the learning rate of the network as 0.001 and it was reduced gradually by a gamma factor of 0.5 every 20 epochs. All models were trained throughout 40 epochs with batch size 32 and implemented in Python v.3.7 on a platform containing a 2.0 GHz Intel Xeon CPU and NVIDA Tesla T4 13 GB GPU. Algorithm 1 and Algorithm 2 describes the training and testing algorithm of the CNN models and the pseudocode for the overall proposed framework, respectively. In the following subsections, the pre-processing, the input data generation, and CNN models are described in more detail. The result of each operation is statistically proven and graphically shown.
4.1 Pre-processing stage
For increasing the productivity and stability of neural networks, image pre-processing is generally considered an indispensable procedure for image-based systems such as CNN. However, it is a complicated and time-consuming procedure, which has to extract the most appropriate features for successful detection. Figure 5 illustrates the applied image-processing techniques used in this work, such as cropping, histogram equalization, and denoising.
In order to produce reliable results and to alleviate the overall computational effort, it is important to select the most effective pre-processing tasks on the available data. It should be observed that an image carries not only spatial information but also information about the interpolated noisy pixels, which causes significant performance degradation or poor learning conditions. Having a large number of noise-free, quality data is an important factor for CNN models to be able to learn effectively without overfitting and to increase the performance of the CNN model considerably [73].
4.2 Data generation
Data augmentation is a frequently preferred data balancing technique in CNN-based methods used to achieve reliable satisfactory results. Although it provides some technical benefits such as increasing the prediction accuracy of the model and reducing overfitting and underfitting, feeding the network with unrealistic data can introduce unnecessary information into the learning process. It also involves additional memory, transformation computation costs, and additional training time. Therefore, the use of data augmentation is limited in this study. In determining the characteristics of data augmentation, the selection of appropriate parameters involves a series of processes, which improve the resolution quality of training data so that steady deep networks can be constructed. After the original datasets were separated into training, validation, and testing, all MR images were augmented t to allow the CNN model to learn the invariant features. These features ultimately improved the model's performance and robustness, in addition to preventing the possibility of overfitting and underfitting. The selected augmentation techniques are rotation, zooming, horizontal flipping, vertical flipping, and height shifting for geometric transformation invariance. In Table 1, to produce better test accuracy rates, the existing dataset has been expanded with the given initial parameters.
The augmented training dataset for tumor detection consists of 9342 images of tumor presence and 8649 images of healthy brain. On the other hand, 9252, 4554, and 6111 new images were generated of the glioma, meningioma, and pituitary tumor types. The statistics of MRI datasets for CNN models are given in Table 2.
4.3 Experiments on EfficientNetv2 versions
The EfficientNetv2 family has 3 types of scales, small (s), medium (m), and large (l). In this study, it was first used to construct a brain tumor diagnosis system. Due to the parameter size of the EfficientNetv2l model being larger than the EfficientNetv2s and EfficientNet2m, it consumed the computer’s GPU resources at an extreme level. As given in Table 3, the size of the trainable parameters used by the models was 20.18 M, 52.86 M, and 117.23 M for s, m, and l, respectively.
As seen in the table, while the pre-trained EfficientNetv2l achieved the best accuracy scores by using learned parameters to the limit of the computer’s computational capability, the training time was much longer when compared to the others (27.68 min with augmentation). It was observed that the increase in model depth increased the training time significantly. The EfficientNetv2s has a fast convergence speed (11.15 min with augmentation) and stable training procedure with a satisfactory test accuracy score of 97.36% on the augmented dataset. Consequently, the EfficientNetv2 can adaptively adjust regularization (e.g. data augmentation) through image size and improve the training procedure using progressive learning. Therefore, when the depth of the model increases, the inputs are resized to 128 × 128 pixels to reduce computer memory consumption. The outputs of the tumor diagnosis systems were evaluated before and after data augmentation, which both improved the overall accuracy, learned invariant features, and reduced overfitting. With the above training strategy, the goal of each model was achieved, while the joint Ranger optimizer minimized the prediction loss.
4.4 Classification performance of optimizers
In the second experiment, we analyzed the competence of optimization algorithms on CNN performance and measured their variability and reliability in automatic classification. Meanwhile, using the Ranger optimizer, instead of the default optimizers, meant the CNN model could avoid overfitting problems and work to ensure a superior classification performance. To evaluate the optimizer, we compared the CNN using Cohen’s Kappa coefficient [18] and Hamming Loss metrics [23] for the 4 optimizers previously mentioned in Section 3.3. The detailed results of each optimizer are given in Table 4. The selected evaluation metrics are expressed in Eq. 1 to Eq. 7 [30].
In these equations, TP, TN, FP, and FN denote the true-positive, true-negative, false-positive, and false-negative values, respectively. Ranger showed notable improvements of 0.9770 ± 0.0094 Cohen’s Kappa, 0.0147 ± 0.0012 Hamming Loss, and 98.60% ± 0.14 in test accuracy compared to the SGD, RMSprop, and Adam, respectively. According to the training time, the SGD achieved the fastest convergence at 32.29 min and the Ranger converged slower than the other optimizers at 42.28 min.
4.5 Comparison with current CNNs
To assess the classification potential of the brain tumor diagnosis system among different CNN models, five types of CNN networks were trained and tested. The values of the metrics containing accuracy, precision, recall, and F1-score were calculated and expressed as mean ± standard deviation in C-category classification. Table 5 compares the various CNN model classification performances found in the 3-tumor type classification of independent test data as micro and macro statistics.
As Table 5 shows, the average training time of every operation related to EfficientNetv2s + Ranger was 41.99 min with a standard deviation of 0.57 over 40 epoch. When completing the same operation the training time for the ResNet18, ResNet200d, and Inception V4 was 12.1, 102.4, and 54.5 min, respectively. The EfficientNetv2s + Ranger model showed a more superior performance than the other CNNs and achieved 99.85% test accuracy, without significantly increasing parameter size and computational cost in repeated tests. As seen in the table, the ResNet18 and InceptionV4 have lower accuracy values than the other CNNs. The ResNet18 has significantly reduced the number of parameters and has a satisfactory performance compared to the InceptionV4. While the ResNet200d is the largest model, the micro and macro scores show that at high training times it achieved almost the same accuracy as the other CNNS. These results indicate that the improved model enabled effective edge and texture learning of tumor characteristics through the use of accuracy and micro and macro statistical metrics. To further confirm the generalization performance of our system, the Receiver Operating Characteristic (ROC) curve and AUC values were also calculated for all models, which are shown in Fig. 6. The ROC curves for our improved model were slightly above other models, with an AUC value 0.9985.
Confusion matrices are presented graphically to identify the discordance between the 3 classes and the CNN models in Fig. 7. While the EfficientNetv2 + Ranger model for glioma tumors outperformed the other models, the Inceptionv4 made an error in predicting only two samples for the meningioma tumor. The ResNet200d appeared to better separate for a pituitary tumor.
4.6 Statistical comparisons
In this section, we used the Cochran Q test statistical method [17] to provide a comprehensive level of type I error and to build pairwise comparisons. It requires that there be only a binary matrix and that there be more than two classifiers of the same size. Figure 8 illustrates the binary comparisons between the models through the Cochran Q test, corresponding to the results presented in Table 5. Assuming that we chose a significance level of α = 0.05, most models in pairwise comparisons did not yield a statistically significant difference except for CV4. The EfficientNetv2 + Ranger showed a statistically significant difference according to both ResNet18 and Inception v4. This indicates that the clear difference may have been caused by the dataset.
Statistical comparisons between different CNN models according to the outputs of fivefold CV. Each mini square shows that the corresponding model was pairwise compared using the Cochran Q test (p > 0.05) to indicate whether there is a statistical difference between them. Blue specifies a statistical difference, orange specifies no statistical difference, and gray specifies the results are not appropriate
5 Results and discussion
In the first experiment, we compared three versions of the EfficientNetv2 family. The dataset with raw images was split into three sets with training, validation, and test. Next, all experiments were conducted on pre-processed augmented and non-augmented images. The hyperparameters were left with default settings, i.e., the learning rate was equal to 0.001, and gamma was set to 0.5 depreciate. All models were run on a fivefold cross-validation set and each training time was noted separately. The accuracy of the models was higher after the data was augmented. In all models, the training time of the models after augmentation was longer than the training time of the model before augmentation. The EfficientNetv2l achieved the highest accuracy of 97.72% using augmentation with the highest training time in the test set as reported in Table 3.
In the second experiment, SGD, RMSprop, Adam, and Ranger optimizers were used on the EfficientNetv2s architecture, respectively. Among these four optimizers, SGD showed poor convergence with 97.36% test accuracy. The Ranger showed slightly better performance using an adaptive momentum based on variance with 98.60% test accuracy, 0.9770 Cohen’s Kappa and 0.0147 Hamming Loss values for the test set as given in Table 4. However, the training time for the network was the longest.
In the third experiment, according to the evaluated and analyzed results using micro and macro metrics, the improved CNN model performed better than the selected state-of-the-art CNN models in diagnosing tumor diseases. These data are presented in Table 5. The ResNet18 and InceptionV4 performed similarly in the classification of test data with 99.62% and 99.69%, respectively. Although the ResNet200d architecture gave the closest scores to what we had obtained, it had the longest training time of 102.48 min due to its deep network structure.
The feature maps of the internal layers extracted from block0 to block3 are illustrated in Fig. 9 to further clarify the learning details of the EfficientNetv2s + Ranger and EfficientNetv2s network with the default optimizer Adam. In the first block, when we compared them for a glioma test sample, we observed that EfficientNetv2s + Ranger had better preserved edges (squared with red boxes) and focused gradients in the tumor region. It is also possible to distinguish this situation from the skull edges. However, EfficientNetv2s + Adam spread the edge details in gradient results. In the next blocks, it can be deduced that final models suppressed weak characterizations such as homogeneous regions in the layer outputs.
Table 6 presents the dataset, the developed environment, a brief explanation, and the accuracy rates of the proposed system with other methods that bring different solutions to the problem. [14] tried to estimate the Isocitrate Dehydrogenase (IDH) mutation status of gliomas from MRI by conducting a residual CNN to preoperative radiographic data. The test accuracy of their model achieved 85.7%. One of the challenges in training CNN networks is that they require a large quantity of training data. In their study, a small amount of data sets from three different private hospitals were used and data augmentation was applied to avoid the model overfitting and underfitting. ELM is another type of learning technique built from one or more layers of hidden nodes. In [57], another CNN model consisting of some intermediate layers was studied to both normalize the data and create an effective mechanism.
The classification accuracy obtained by the KE-CNN model was 93%. [11] explored the performance of the three distinct deep CNN architectures trained on malignant intracranial MR images to discriminate between High-Grade Glioma (HGG) and Low-Grade Glioma (LGG). The VolumeNet was accomplished using the 3D volumetric dataset instead of 2D and while it has greatly improved findings with 97% classification accuracy, the large size of the model is a disadvantage. [45], inspired by ResNet34 architecture, designed a CNN model which mainly consists of a global pooling layer in addition to all-convolutional layers. They called their network G-ResNet and in their approach, the accuracy rate was measured as 95% due to the reduced number of parameters. [24] developed a new CNN model combining the typical trait groups to recognize IDH mutations and TERT promoter (pTERT) mutations. Simultaneously, the tumor lesion area was extracted from the image and normalized before they were sent to the AlexNet architecture. The model performance was measured with a final accuracy of 63.1% on a private dataset. Another brain tumor classification method using the Deep CNN (DCNN) architecture was experimented with in [10] using only 22 layers and patches extracted from whole MR images. Assuming that the hardware resources were limited, their shallow model can be appropriate in the case of narrow network bandwidths such as mobile phones, conventional PCs, or short response time. Meanwhile, in [46], a GAN-based model using ResNet50 on public and private datasets was implemented to further exhibit the feasibility and productivity of diagnosing IDH status of glioma tumors. The authors emphasized that IDH is a significant biomarker for glioma and ultimately results in better test accuracy of 88%. A wealth of research has considered modeling the detection and identification of brain tumors and therefore [51] prepared noteworthy prospective research that tries to solve this issue based on recent AI. In this survey, the accuracy rates of the two models generated by [61] and [15] based on VGGNet were 93% and 94%, respectively. The approaches given in [7, 22], and [25] classify t1-weighted contrast-enhanced MR images. [22] discusses a multi-scale deep architecture that allows application at three spatial scales through different procedures. However, this idea is self-diagnostic after segmentation and it causes the model to be more computationally complex and impractical for medical applications due to the increased amount of errors. Rather than handling segmentation, [7] showed that tumor type can be estimated from optimized GoogleNet and ResNet101 with transfer learning. They attained the highest performance of 99.33% in tumor detection and 95.65% in tumor diagnosis. More recently, ensemble learning methods for diagnosing brain tumors have been able to deal with more reliable feature extraction from MR images in the case that small amounts of brain tumor data are available. First, brain tumor recognition was performed using a modified InceptionResNet-pretrained model in [25]. In the second phase, in the case when a tumor lesion was detected, the tumor type was specified using the combination of InceptionResNet and Random Forest Tree (RFT). The last line in the table gives the accuracy of our methodology. Among all of the methods for tumor detection, the test accuracy of our method gave almost the same result compared to [25] at 99%, the EfficientNetv2 + Ranger yielded the best performance for tumor classification at 98.85%.
Although the proposed CNN-based approach can be used as an alternative to the existing methods of brain tumor diagnosis, there are some limitations to its performance. These depend on, among other factors, the dataset used in the training, the limitation of MRI scanning, and the deep learning methodology involved in training such systems. Two publicly available datasets were used in this study, thus some clinical information about patients was limited. To improve the robustness of the CNN models, future multicenter studies with more detailed clinical data including patient datasets are required. Additionally, because the radiological features of primary tumors, metastases, and central nervous system cysts are similar, conventional MRIs are not precisely reliable to discriminate between them. Therefore, additional examinations such as computer tomography and biopsy are required for the exact diagnostics. CNN-based methods also have structural limitations. For example, some problems such as misclassification may be encountered in the application of CNN models, thus they may be ineffective in making the correct diagnosis of brain tumors and may be expensive in terms of computational costs.
6 Conclusions
In cases with high or weak contrast in MRI brain images, there may be uncertainty when deciding the presence of a tumor. Computer-aided diagnostic systems can support clinicians and radiologists in distinguishing between tumors and healthy cells. With radiological imaging, the presence of cancer and similar diseases is observed, and information about its size and location is obtained. With the help of AI-based solutions, reliable clarification of the diagnosis of tumor tissues with high similarity can be achieved using algorithm innovation. We presented an end-to-end CNN-based brain tumor classification system that represents a powerful approach for meningioma, glioma, and pituitary tumors based on MRI in neurosurgery clinics and demonstrated the following findings:
-
We conducted the EfficientNetv2 + Ranger model, which had not been previously considered in the literature, and we achieved 99% test accuracy in tumor detection and 99.85% test accuracy in predicting meningioma, glioma, and pituitary tumors.
-
In CNN networks, applying extensive pre-processing on the raw dataset exhibited significantly improved learning capabilities.
-
The Ranger algorithm provides robust and stable training throughout the training period in different CNN architectures by reducing the variance.
-
By performing a wide range of experiments on tumor datasets, we examined recent CNN models and optimizers and emphasized convenient choices for recognition in smart healthcare.
This study stated that AI-based methods can support radiologists in validating their initial scans of brain tumors for multi-classification purposes. Unfortunately, for the indicated contributions the available datasets were obtained retrospectively by the dataset provider and the clinical diagnoses of the patients were not exactly confirmed. In a case where the patient has different symptoms, the detection of a brain tumor can change the diagnosis and treatment planning for clinicians and medical pathologists. In addition, the use of CNN for tumor diagnosis requires the identification of features such as shape, size, location, and extensive abnormalities in various directions of tumors, rather than the handcrafted features. Another challenge is that in this problem, it should be noted that the implementation of CNN models can be ineffective and computationally expensive.
Despite these adverse conditions, the cutting-edge AI-based solutions have made great progress in identifying cancers in recent years. In this way, the proposed method encourages the development of current patient diagnosis techniques by including CNN-based techniques in the use of the health profession. This study paves the way for incorporating CNNs into medicine by significantly increasing the rate of cancer recognition.
We will refine the diagnostic output by considering the 3D spatial information among the brain MR slices using a lightweight CNN architecture, inspired by some recent studies using radiomic analyzes [37, 42, 65]. We also plan to collect information on the clinical features of a custom dataset so that we can use the data from patients suffering from brain tumors in CNN networks as complementary discriminative features in our future works.
References
Abdelali HA, Derrouz H, Zennayi Y, Thami ROH, Bourzeix F (2021) Multiple Hypothesis Detection and Tracking Using Deep Learning for Video Traffic Surveillance. IEEE Access 9:164282–164291
Abuhamad M, Abuhmed T, Mohaisen D, Nyang D (2020) Deep-Learning-Based Implicit Continuous Authentication Using Smartphone Sensors. IEEE Internet Things J 7:5008–5020
Ahmad I, Shin S (2022) A perceptual encryption-based image communication system for deep learning-based tuberculosis diagnosis using healthcare cloud services. Electronics 11:(16). https://doi.org/10.3390/electronics11162514
Alhassan AM, Zainon WMNW (2021) Brain tumor classification in magnetic resonance image using hard swish-based RELU activation function-convolutional neural network. Neural Comput Appl 33:9075–9087
Ali SI, Ebrahimi SS, Khurram M, Qadri SI (2021) Real-Time Face Mask Detection in Deep Learning using Convolution Neural Network, 2021 10th IEEE International Conference on Communication Systems and Network Technologies (CSNT). 639–642
Alqudah AM, Alquraan H, Abu-Qasmieh I, Alqudah A, Al-Sharu W (2020) Brain Tumor Classification Using Deep Learning Technique - A Comparison between Cropped, Uncropped, and Segmented Lesion Images with Different Sizes, ArXiv, abs/2001.08844
Anjum S, Hussain L, Ali M, Alkinani H, Aziz W, Gheller S, Abbasi A, Marchal A, Suresh H, Duong T (2022) Detecting brain tumors using deep learning convolutional neural network with transfer learning approach. Int J Imaging Syst Technol 32. https://doi.org/10.1002/ima.22641
Arunkumar N, Mohammed MA, Abd Ghani MK, Ibrahim DA, Abdulhay E, Ramirez-Gonzalez G, de Albuquerque VHC (2019) K-Means clustering and neural network for object detecting and identifying abnormality of brain tumor. Soft Comput 23:9083–9096
Arunkumar N, Mohammed M, Mostafa S, Ibrahim D, Rodrigues J, Albuquerque VHC (2018) Fully automatic model‐based segmentation and classification approach for MRI brain tumor using artificial neural networks. Concurr Comput Pract Exp 32. https://doi.org/10.1002/cpe.4962
Badza M, Barjaktarovic M (2020) Classification of Brain Tumors from MRI Images Using a Convolutional Neural Network. Appl Sci 10:1999
Banerjee S, Mitra S, Masulli F, Rovetta S (2019) Brain Tumor Detection and Classification from Multi-sequence MRI: Study Using ConvNets: 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Revised Selected Papers, Part I. 170-179
Brown CE, Bucktrout S, Butterfield LH, Futer O, Galanis E, Hormigo A, Lim M, Okada H, Prins R, Marr SS, Tanner K (2022) The future of cancer immunotherapy for brain tumors: a collaborative workshop. J Transl Med 20:236
Cabrera C, Cervantes D, Muñoz F, Hirata G, Juárez P, Flores D-L (2022) Classifying nanostructured and heterogeneous materials from transmission electron microscopy images using convolutional neural networks. Neural Comput Appl 34:11035–11047
Chang K, Bai H, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis V, Senders J, Boaro A, Beers A, Zhang B, Capellini A, Liao W, Shen Q, Li X, Xiao B, Cryan J, Ramkissoon S, Ramkissoon L, Kalpathy-Cramer J (2017) Residual convolutional neural network for determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res 24(5):1073–1081. https://doi.org/10.1158/1078-0432.CCR-17-2236
Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, Feng Q (2015) Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 10. https://doi.org/10.1371/journal.pone.0140381
Cheng J (2017) T1-weighted contrast-enhanced brain tumor dataset. Figshare. https://figshare.com/articles/dataset/brain_tumor_dataset/1512427. Accessed 01.09.2022
Cochran WG (1950) The Comparison of Percentages in Matched Samples. Biometrika 37:256–266
Cohen J (1960) A Coefficient of Agreement for Nominal Scales. Educ Psychol Measur 20:37–46
Cruzulloa C, Krus A, Barrientos A, Cerro JD, Valero C (2022) Robotic Fertilization in Strip Cropping using a CNN Vegetables Detection-Characterization Method. Comput Electron Agric 193:106684
Dabral I, Singh M, Kumar K (2021) Cancer detection using convolutional neural network, pp 290–298. https://doi.org/10.1007/978-3-030-67187-7_30
Darbari A, Kumar K, Darbari S, Patil P (2021) Requirement of artificial intelligence technology awareness for thoracic surgeons. Cardiothorac Surg 29:13
Díaz-Pernas F, Martínez Zarzuela M, Antón-Rodríguez M, González-Ortega D (2021) A Deep Learning Approach for Brain Tumor Classification and Segmentation Using a Multiscale Convolutional Neural Network. Healthcare 9:153
Frank E, Hall M (2001) A Simple Approach to Ordinal Classification. In: De Raedt L, Flach P (eds) Machine Learning: ECML 2001. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 145–156
Fukuma R, Yanagisawa T, Kinoshita M, Shinozaki T, Arita H, Kawaguchi A, Takahashi M, Narita Y, Terakawa Y, Tsuyuguchi N, Okita Y, Nonaka M, Moriuchi S, Takagaki M, Fujimoto Y, Fukai J, Izumoto S, Ishibashi K, Nakajima Y, Haruhiko K (2019) Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network. Sci Rep 9:20311
Gupta RK, Bharti S, Kunhare N, Sahu Y, Pathik N (2022) Brain tumor detection and classification using cycle generative adversarial networks. Interdiscip Sci Comput Life Sci 14(2):485–502. https://doi.org/10.1007/s12539-022-00502-6
Hamada A (2020) Brain Tumor Detection (BR35H) Dataset, Kaggle
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H (2017) Brain tumor segmentation with Deep Neural Networks. Med Image Anal 35:18–31
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 27-30 June 2016, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Heidari H, Chalechale A (2022) Biometric authentication using a deep learning approach based on different level fusion of finger knuckle print and fingernail. Expert Syst Appl 191:116278
Hossin M, Sulaiman MN (2015) A review on evaluation metrics for data classification evaluations. Int J Data Min Knowl Manag Process 5:01–11. https://doi.org/10.5121/ijdkp.2015.5201
Howard A, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H (2017) MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications
Huang M-L, Liao Y-C (2022) A lightweight CNN-based network on COVID-19 detection using X-ray and CT images. Comput Biol Med 146:105604
Huang G, Liu Z, van der Maaten L, Weinberger K (2017) Densely Connected Convolutional Networks
Husham S, Mustapha A, Mostafa S, Al-obaidi M, Mohammed M, George ST (2022) Comparative Analysis between Active Contour and Otsu Thresholding Segmentation Algorithms in Segmenting Brain Tumor Magnetic Resonance Imaging
Ji Z, Zhao Y, Pang Y, Li X, Han J (2021) Deep Attentive Video Summarization With Distribution Consistency Learning. IEEE Trans Neural Netw Learning Syst 32:1765–1775
Jia S, Jiang S, Lin Z, Li N, Xu M, Yu S (2021) A survey: Deep learning for hyperspectral image classification with few labeled samples. Neurocomputing 448:179–204
Kan Y, Dong D, Zhang Y, Jiang W, Zhao N, Han L, Fang M, Zang Y, Hu C, Tian J, Li C, Luo Y (2018) Radiomic signature as a predictive factor for lymph node metastasis in early‐stage cervical cancer. J Magn Reson Imaging 49. https://doi.org/10.1002/jmri.26209
Kaur T, Gandhi TK (2019) Automated Brain Image Classification Based on VGG-16 and Transfer Learning, 2019 International Conference on Information Technology (ICIT) pp. 94–98
Kingma D, Ba J (2014) Adam: A Method for Stochastic Optimization, International Conference on Learning Representations
Kumar BV, Kumar P (2019) Brain tumor MRI segmentation and classification using ensemble classifier. Int J Recent Technol Eng 8:244–252
Kumari S, Singh M, Kumar K. Prediction of Liver Disease Using Grouping of Machine Learning Classifiers, in: M. Tripathi, S. Upadhyaya (Eds.) Conference Proceedings of ICDLAIR2019, Springer International Publishing, Cham, 2021, pp. 339–349
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S (2017) Radiomics: the bridge between medical imaging and personalized medicine, Nature Reviews. Clin Oncol 14:749–762
Lim S-C, Huh J-H, Hong S-H, Park C-Y, Kim J-C (2022) Solar power forecasting using CNN-LSTM hybrid model. Energies 15:(21). https://doi.org/10.3390/en15218233
Liu L, Jiang H, He P, Chen W, Liu X, Gao J, Han J (2019) On the variance of the adaptive learning rate and beyond. ArXiv abs/1908.03265
Liu D, Liu Y, Dong L (2019) G-ResNet: Improved ResNet for Brain Tumor Classification. In: Gedeon T, Wong KW, Lee M (eds) Neural Information Processing. Springer International Publishing, Cham, pp 535–545
Liu S, Shah Z, Sav A, Russo C, Berkovsky S, Qian Y, Coiera E, Di Ieva A (2020) Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci Rep 10:7733
Mansour RF, Escorcia-Gutierrez J, Gamarra M, Villanueva JA, Leal N (2021) Intelligent video anomaly detection and classification using faster RCNN with deep reinforcement learning model. Image Vis Comput 112:104229
Mathada R, Vidya CS, Patel S, Suma MN (2022) Arachnoid cyst: a solitary intracranial cyst in the occipital lobe of the cerebrum in a cadaver. J Clin Diagn Res 16. https://doi.org/10.7860/JCDR/2022/51377.15889
Maurovich-Horvat P (2019) 2 Steps Forward, 1 Step Back. JACC Cardiovasc Imaging 12:1377–1378
Mekruksavanich S, Jitpattanakul A (2021) Deep Learning Approaches for Continuous Authentication Based on Activity Patterns Using Mobile Sensing. Sensors 21:7519
Muhammad K, Khan S, Ser JD, Albuquerque VHCD (2021) Deep Learning for Multigrade Brain Tumor Classification in Smart Healthcare Systems: A Prospective Survey. IEEE Trans Neural Netw Learning Syst 32:507–522
Negi A, Chauhan P, Kumar K, Rajput RS (2020) Face Mask Detection Classifier and Model Pruning with Keras-Surgeon, 2020 5th IEEE International Conference on Recent Advances and Innovations in Engineering (ICRAIE). 1–6
Negi A, Kumar K (2021) Classification and detection of citrus diseases using deep learning (1st edn). Chapman and Hall/CRC, p 23
Negi A, Kumar K (2022) Chapter 1 - AI-based implementation of decisive technology for prevention and fight with COVID-19, in: R.C. Poonia, B. Agarwal, S. Kumar, M.S. Khan, G. Marques, J. Nayak (Eds.) Cyber-Physical Systems, Academic Press. 1–14
Negi A, Kumar K, Chauhan P (2021) Deep learning-based image classifier for malaria cell detection. In: Machine Learning for Healthcare Applications, pp 187–197. https://doi.org/10.1002/9781119792611.ch12
Negi A, Kumar K, Chauhan P (2021) Deep neural network-based multi-class image classification for plant diseases. In: Agricultural Informatics, pp 117–129. https://doi.org/10.1002/9781119769231.ch6
Pashaei A, Sajedi H, Jazayeri N (2018) Brain Tumor Classification via Convolutional Neural Network and Extreme Learning Machines, 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE) pp. 314–319
Pravitasari AA, Iriawan N, Almuhayar M, Azmi T, Irhamah I, Fithriasari K, Purnami SW, Ferriastuti W (2020) UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation. TELKOMNIKA (Telecommunication, Computing, Electronics and Control) 18(3):1310–1318. https://doi.org/10.12928/TELKOMNIKA.v18i3.14753
Rajinikanth V, Kadry S, Damaševičius R, Sujitha RA, Balaji G, Mohammed MA (2022) Glioma/Glioblastoma Detection in Brain MRI using Pre-trained Deep-Learning Scheme, 2022 Third International Conference on Intelligent Computing Instrumentation and Control Technologies (ICICICT). 987–990
Sadad T, Rehman A, Munir A, Saba T, Tariq U, Ayesha N, Abbasi R (2021) Brain tumor detection and multi-classification using advanced deep learning techniques. Microsc Res Tech 84:1296–1308
Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW (2019) Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci 30:174–182
Sarker MMK, Akram F, Alsharid M, Singh VK, Yasrab R, Elyan E (2023) Efficient breast cancer classification network with dual squeeze and excitation in histopathological images. Diagnostics 13(1):103
Sifre L, Mallat S (2014) Rigid-motion scattering for texture classification. ArXiv abs/1403.1687
Simonyan K, Zisserman A (2014) Very Deep Convolutional Networks for Large-Scale Image Recognition, arXiv 1409.1556
Stadlbauer A, Marhold F, Oberndorfer S, Heinz G, Buchfelder M, Kinfe T, Meyer-Bäse A (2022) Radiophysiomics: Brain Tumors Classification by Machine Learning and Physiological MRI Data. Cancers 14:2363
Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions, 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) pp. 1–9
Tan M, Le Q (2021) EfficientNetV2: smaller models and faster training. ArXiv abs/2104.00298
Tong Q, Liang G, Bi J (2022) Calibrating the adaptive learning rate to improve convergence of ADAM. Neurocomputing 481:333–356
Ur Rahman A, Saeed M, Saeed MH, Zebari DA, Albahar M, Abdulkareem KH, Al-Waisy AS, Mohammed MA (2023) A framework for susceptibility analysis of brain tumours based on uncertain analytical cum algorithmic modeling. Bioengineering 10(2):147
Wahid A, Breslin JG, Intizar MA (2022) Prediction of machine failure in industry 4.0: a hybrid CNN-LSTM framework. Appl Sci 12(9):4221
Wong KCL, Syeda-Mahmood T, Moradi M (2018) Building medical image classifiers with very limited data using segmentation networks. Med Image Anal 49:105–116
Yu Y, Cao Z, Liu Z, Geng W, Yu J, Zhang W (2022) A Two-Stream CNN With Simultaneous Detection and Segmentation for Robotic Grasping. IEEE Trans Syst Man Cybern Syst 52:1167–1181
Zhang Y-D, Dong Z, Chen X, Jia W, Du S, Muhammad K, Wang S-H (2019) Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed Tools Appl 78:3613–3632
Zhang P, Yang L, Li D (2020) EfficientNet-B4-Ranger: A novel method for greenhouse cucumber disease recognition under natural complex environment. Comput Electron Agric 176:105652
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Paper presented at the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)
Acknowledgements
This research did not receive any funding from any organization in the public or private sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The author (s) declare there is no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Anagun, Y. Smart brain tumor diagnosis system utilizing deep convolutional neural networks. Multimed Tools Appl 82, 44527–44553 (2023). https://doi.org/10.1007/s11042-023-15422-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15422-w