Abstract
There is a broad range of novel Coronaviruses (CoV) such as the common cold, cough, and severe lung infections. The mutation of this virus, which originally started as COVID-19 in Wuhan, China, has continued the rapid spread globally. As the mutated form of this virus spreads across the world, testing and screening procedures of patients have become tedious for healthcare departments in largely populated countries such as India. To diagnose COVID-19 pneumonia by radiological methods, high-resolution computed tomography (CT) of the chest has been considered the most precise method of examination. The use of modern artificial intelligence (AI) techniques on chest high-resolution computed tomography (HRCT) images can help to detect the disease, especially in remote areas with a lack of specialized physicians. This article presents a novel metaheuristic algorithm for automatic COVID-19 detection using a least square support vector machine (LSSVM) classifier for three classes namely normal, COVID, and pneumonia. The proposed model results in a classification accuracy of 87.2% and an F1-score of 86.3% for multiclass classifications from simulations. The analysis of information transfer rate (ITR) revealed that the modified quantum-based marine predators algorithm (Mq-MPA) feature selection algorithm reduces the classification time of LSSVM by 23% when compared to the deep learning models.
Similar content being viewed by others
1 Introduction
COVID-19 viruses contain single-stranded (positive-sense) Ribonucleic Acid (RNA) with a nucleoprotein within a capsid consisting of matrix protein. According to Mohanty et al., 2020 [32], the first case of Coronavirus (COVID-19) was reported on December 1, 2019. It was later discovered that a new coronavirus, later named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) may have originated in animals and evolved (mutated) to cause illness in humans. Three people in the Wuhan area in China with pneumonia were identified as the first to carry SARS-CoV-2 [17]. COVID-19 has infected 257 million people since November 2021, and it has led to the deaths of 5.15 million people. It was declared a global pandemic by the World Health Organization (WHO) [17]. There has been widespread COVID-19 infection in 213 countries, with cases exceeding 0.1 million in countries such as the United States, Italy, India, China, the United Kingdom, Spain, Brazil, and Russia [37].
The initial study on the spread of COVID-19 reported that the virus is highly infectious and spreads mainly due to droplets via coughing or sneezing [16]. Many countries conducted awareness because of minimizing the spread of the virus by closing international borders and travels as the cure or vaccine was unavailable. All the counties were focused on screening the people to spot the infection of the virus and allowing the patients to isolate themselves under medical supervision or self-isolation based on the patient’s condition [39]. Only one test was available for screening: reverse transcription-polymerase chain reaction (RT-PCR). However, RT-PCR involves an invasive method of collecting the sample(s), which is usually taken by swabbing the patient’s nose or throat [46]. The disadvantages of RT-PCR tests include high false results, low quality of the sample, and delay in obtaining the results [26]. A shortage of medical professionals is causing a delay in the collection and performance of RT-PCR tests for COVID-19 because it requires extensive training. Further, the false-negative results of the test present a new challenge of further spread of the virus as affected people started visiting the test centers. It further complicates the situation of the patient from obtaining appropriate treatment.
It has been reported that some patients with severe infections develop respiratory failure and need intensive care within 48 hours of being infected by the COVID-19 virus [28]. For such patients with severe infections, mechanical ventilation should be used immediately for intensive care. This implies that there is an acute need to precisely detect the virus and use rapid and reliable diagnostic tools as opposed to only RT-PCR [22]. As a result, computed tomography (CT) imaging is a non-invasive alternative method for finding COVID-19 pneumonia patients by detecting the typical radiographic features [29].
The absence of expert radiologists to analyze and report the CT images for diagnosis is like the manual application of magnetic resonance imaging (MRI) for diagnosis. The result can be erroneous decisions based on the lack of expert radiologists. The use of Machine Learning algorithms to automate the process of CT image analysis has been proposed in the literature to detect COVID-19 [14]. Compared to RT-PCR tests, CT has a sensitivity of 98% for the detection of COVID-19 infection, while RT-PCR has only a 71% sensitivity [27]. The automation of CT image analysis may also assist in the avoidance of medicinal delays and errors introduced due to fatigue-related readings of images.
In this article, we propose a model, which has the following main stages: CT image pre-processing, lung infected region segmentation, texture analysis for feature extraction, selection of optimal features using the proposed metaheuristic algorithm, and multiclass classification using LSSVM [44] and convolutional neural networks (CNN). At first, the input CT image is pre-processed by contrast limited adaptive histogram equalization (CLAHE) for the selected green channel of the image. In the next step, lung infected region segmentation using multilevel thresholding is performed. Further, texture image analysis is performed using gray-level co-occurrence matrix (GLCM) and Gabor filtering techniques. In the following phase, Shannon’s and Kapur’s entropies are extracted to know how much energy and entropy are present in the texture. To shrink the lengthy feature vector, we propose a unique feature selection algorithm with a pseudo-random number generator meta-heuristic approach called the modified Mq-MPA. By utilizing a novel evolutionary computing-based feature selection algorithm, our proposed model classifies the COVID-19-infected patients automatically based on CT images. Due to this automated diagnostic tool, medical professionals will be able to speed up the diagnosis process, resulting in a reduction in their workload. Further, the present literature on COVID-19 classification using CT scan images focuses mainly on deep learning (DL) algorithms [20]. In our study, we propose a novel assessment of the COVID-19 CT image diagnosis by using the image processing algorithms, extraction of relevant features, optimal feature selection algorithm, and multi-class LSSVM. After converting the green channel of each RGB channel into a metaheuristic algorithm, a selection of the best features is obtained from the chest imaging CT scan. Comparing the proposed method to the DL method directly, the use of a feature selection algorithm contributes to better results. As demonstrated in [35], researchers often incorporate empirical wavelet transform (EWT) for the purpose of feature extraction. Lastly, our proposed technique is then compared with the DL enabled classification using transfer learning.
Overall, the contribution of the work presented in this article is as follows:
-
1)
COVID-19 screening using chest CT scan images for classifying the virus infection as positive, negative, or pneumonia is presented using an ML algorithm.
-
2)
Proposed the combination of advanced image processing, feature extraction, and selection algorithms are implemented to overcome the drawback of RT-PCR by reducing high false-negative values for classification of COVID-19 negative, pneumonia, and positive samples.
-
3)
A model is proposed by extracting only the green channel of images in the R, G, and B regions of the CT scans, and then determining features with a metaheuristic algorithm based on the local binary pattern (LBP) and entropy feature extraction.
-
4)
The feature selection is based on a novel Mq-MPA algorithm which is a metaheuristic algorithm and resulted in enhanced accuracy of the LS-SVM classifier. In addition, the proposed model is evaluated by comparing it with the existing approaches.
-
5)
A comparison of the LS-SVM and CNN classifiers based on computational complexity and size of the dataset used in the study is performed given these two parameters being important for embedded system development of a portable COVID-19 detection system.
The remaining manuscript is structured as follows: Section 2 presents the related work and review. In Section 3, we present details of the proposed method. Section 4 describes the feature extraction and selection algorithms. Section 5 presents results and discussion. Finally, Section 6 concludes the paper.
2 Literature review
In this section, we present a review of significant literature on COVID-19 classification and detection using chest X-ray and CT images. It is to be noted that the focus of this study is to achieve optimum algorithms for the implementation as an embedded device.
2.1 Related work
In [30], the authors analyzed chest X-rays to classify the “binary” and “multiple” classes of COVID-19 using a deep support vector machine (DSVM) and depthwise separable convolution neural network (DWS-CNN). The Gaussian-based filtering of images is proposed in the first stage of DWS-CNN followed by feature extraction and classification using DSVM. With the chest X-ray image dataset, the simulation process of the DWS-CNN model is assessed, and experimental results show a high classification accuracy of 98.54% to 99.06% for binary and multiple class classification, respectively. Further, the authors have also highlighted the use of bio-inspired algorithms for higher results in the future.
Using a combination of the standard reconstruction error and an LSSVM loss function, the authors [12] report a practical chest X-ray classification application. The authors report that the learning features with linear class separability can increase classification accuracy. In the study, on the COVIDGR dataset, a convolutional auto-encoder has been employed, and the improvements reported by the feature vectors, use of feature learners, and conventional classifiers are discussed in the study.
In [7], the authors have addressed a two-class problem using the CNN and the CT image features. The authors have demonstrated an enhancement in the feature representation via a technique of feature combination, which increases the representation power. In addition, a unique way of combining the features and ranking them is presented in the study considering 150 CT images for an SVM classifier with 98.27% accuracy.
The authors [33] have used SVM and logistic regression to classify COVID-19 and non-infected patients by processing the chest X-ray. The histogram of oriented gradients has been demonstrated to show an accuracy of 96% when compared with the logistic regression. It is this study that has motivated our employment of LSSVM for the multi-class problem in the current study.
In [47], the authors studied 1396 lung CT images using a 23-layer CNN and compared the performance with SVM and k-Nearest Neighbors (k-NN). The authors have mentioned 2-fold and 10-fold cross-validation for a texture analysis method, and have used various performance parameters such as accuracy, sensitivity, specificity, F-1 score, and area under the ROC curve (AUC).
The authors [24] have conducted a study on CT images, and have presented a performance comparison of ANN, SVM, and decision tree (DT). Researchers have developed a neuro-fuzzy inference system based on adaptive neuro-fuzzy inferences (ANFIS) which achieves 98.53% accuracy and a testing time of 0.02 seconds. This study motivated our analysis of the computation time of various algorithms for COVID-19 CT images. Also, we have focused on the implementation of the system used in this study for the low execution time of embedded system demands on lightweight processors.
In [23], a novel technique of splitting the chest X-ray image into several distinct areas, called a multi-level threshold, is used to extract the image features and the classification of COVID-19. A method for identifying different X-ray thresholds and splitting images by corresponding intensities into areas with one background and a variety of objects has been proposed in different studies. The use of the SVM algorithm has resulted in 97.48% of classification accuracy.
Researchers have compared five different DL models to detect COVID-19 infection using chest CT images [31]. The models are VGGNet19, AlexNet, VGGNet16, ResNet50, and GoogleNet. Classical data augmentation methods coupled with conditional generative adversarial nets (CGAN) appear to improve classification performance across all deep transfer models. In the case of limited chest CT images, the ResNet50 model demonstrated the most appropriate results with 82.91% accuracy.
[21] present a technique for abnormality localization using “pre-trained CNNs” and Phase-3 augmented data in CT scan imaging. These experiments have been carried out using pre-trained models including ResNet50ResNet18, SqueezeNet, and ResNet101.
Authors [4] have compared an aggregate of 15 pre-trained CNN models for COVID-19 CT image classification, as COVID (positive) and COVID (negative). The authors used these architectures and then “fine-tuned” them on the target class. A public dataset of CT scans is used to demonstrate that an ensemble method with majority voting improves recognition performance. Using 10 models, the authors report an accuracy of 85%. They include EfficientNets (B0-B5), NasNetLarge, NasNetMobile, Xception, DenseNet121, ResNext50, and Inception_resnet_v2. In addition, results suggest that using the enhanced performance of five models from deep transfer learning (EffectiveNetB0, EffectiveNetB3, EffectiveNetB5, Inception_resnet_v2, and Xception) leads to improved outcomes than using them individually.
A comprehensive review of steps used in detecting COVID-19 using CT image analysis is presented by [9] emphasizing the impact of computer vision and deep learning. Authors [6] present an alternate approach of using machine learning models to detect COVID-19 using 14 features for six different classifiers and thus comment on the shortage of RT-PCR tests. An accuracy of 84.21% is reported using the technique of machine learning tools. Author [18] propose a modified ResNet50 on a large dataset of images and report 97.7% accuracy on the pre-trained model. Authors [15] proposed a modification in the lung and infection segmentation Steps by using residual block techniques, called Residual U-Net to produce an average specificity of 99.76%.
2.2 Review
Although the literature presents many mechanisms and algorithms to classify COVID-19 images as 2-class and 3-class problems, there exist a few shortcomings of these methods which need to be addressed effectively to enhance the performance of automated detection systems. COVID-19 detection and classification methods have several benefits as well as limitations.
Among all the studies mentioned in Table 1, CNN’s results in high performance [4]. Also, as a rule of thumb, DL is known to perform well when dealing with large datasets such as image classification [21]. However, a major shortcoming of using the DL algorithms in the embedded system scenario is the need to deploy a high-end dedicated processing unit. For the DL algorithms to perform well, they need large datasets and a mechanism for training them in a reasonable amount of time. The final drawback is the complexity of the DL algorithms because they require much more practice than any off-the-shelf classifier.
However, SVMs, which have a large margin interpretation, are efficient for many classification problems with relatively small data sets and lesser noise. In SVMs, the (convex) quadratic programming (QP) problem is used to characterize a classification problem. It is possible to tailor SVMs to obtain a linear set of equations in the dual space using the LS-SVMs [44]. Additionally, binary classifiers with varying output coding schemes are used in Multiclass categorization problems. In addition to regularization controlling the number of effective parameters of the LS-SVM, the choice of the 2-norm alters the sparseness property of SVMs. During the sparse approximation procedure, the support value spectrum can be pruned gradually, and hyper-parameters optimized to enforce sparsity in a second stage. Hence, the above-specified merits can be utilized to develop novel algorithms for COVID-19 classification and detection using the optimized feature selection strategy.
By considering the comparison of the studies mentioned in Table 1, in the proposed study, we use the LS-SVM technique in combination with a bio-inspired algorithm for CT image feature selection. In the literature, authors have employed CNN for CT image classification for COVID-19 classification. The use of a metaheuristic algorithm for feature selection resulting in optimizing the performance of LS-SVM is our main goal of this work. The classifier is implemented as a multi-class classifier using a binary to multi-class approach [5].
3 Lung infected region segmentation
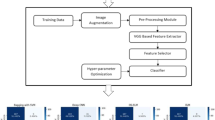
In this section, we describe the proposed methods for image pre-processing and segmentation. By automatically identifying and classifying the variations in the chest CT images as COVID-19 positive, pneumonia, and negative categories, the model is designed to detect and classify the images. A COVID-19 lung CT scan is assessed in this section to see whether it is positive, negative, or Pneumonia. The general block diagram for the system in Fig. 1, shows the process flow behind the operation.
The limitations in the chest CT recording device result in compromised spatial resolutions in most cases of recording the scan. The image pre-processing through image channel selection in the first step ensures lower computational time for further algorithms. Also, for enhancement of contrast, the CLAHE technique [49] is applied to the images. An optimal segmentation and thresholding of the contrast-enhanced image is done using the Marine Predictor (MPA) Algorithm [19] to determine the infected lung region. The algorithm uses the operators of whale optimization and improves the learning phase of the traditional volleyball premier league (VPL) algorithm to employ an image histogram as the input to maximize “Otsu’s function” for computing the optimal threshold required in image segmentation. Following are the steps for extracting texture features from an image and determining its energy, such as LBP and texture energy features (TEF). After segmenting the images for abnormalities, entropies are extracted. Entropy parameters of an image, and Kapur’s and Shannon’s entropy provide this measurement. To optimize the feature length, an innovative feature selection technique based on the meta-heuristic approach called MPA is implemented in the current work. The LS-SVM performs the task of multiclass classification by reducing the problem to a set of binary classes. The optimal feature class for the LS-SVM algorithm is considered to compare it with CNN.
As a preliminary step, we define chest CT images for every stage of the system as follows: the input CT image is represented as IimCT, the pre-processed image is denoted as Iimpre, lung infected region segmented image is denoted as IimLIR, and the final distinctive features of COVID-19, represented by abnormalities such as dilated segments and temporal changes, is denoted as an image Imimseg. The feature segments, extracted from the segmented images, are indicated by Llbpi, Ltxtei and Lenti where i = 1, 2, ⋯, k, and k shows the number of feature samples. Lastly, the selection of feature vectors with high variance is denoted as an optimal feature set Wopt∗Fset where Fset = 1, 2,⋯, Fopt indicates several optimum features.
3.1 Pre-processing of COVID-19 images
A pre-processing stage includes a green channel selection and a CLAHE on the CT images.
-
(i)
A typical input CT image consists of three RGB channels, namely red, green, and blue, which have low contrast due to radiation dose and noise. In our study, we transform the images into green channel images since the green channel has the highest contrast and it can represent image abnormalities.
-
(ii)
CLAHE: In this algorithm, the most frequent intensity value in an image is stretched by spreading the intensity range of the image or by stretching the AHE algorithm. Stretching the intensity values, however, causes the input image to acquire undesired noise and change its natural brightness. CLAHE is an effective method for enhancing the brightness of the image [36].
The algorithm, which improves the contrast of CT images using the CLAHE technique, is as follows:
-
a)
As a result of over-amplifying the histograms, the AHE method introduces noise into the image due to the equally distributed computation of the histograms. However, CLAHE works by splitting the histogram of the input image IimCT at precise values and limits the amplification before calculating the cumulative distributive function (CDF).
-
b)
The intensity values of the histogram are used to extract each contextual region.
-
c)
A threshold parameter is used to clip the histograms as a limit to describe the defined brightness level of the CT image.
-
d)
Each histogram is modified using the selected transformation functions.
-
e)
The selected clip limit defines the change in each histogram with a condition that the selected value does not exceed the clip limit.
The calculation of the CLAHE technique is given in Eq. (1),
where the extreme pixel value is denoted as PCTmax, PCTmin is least pixel parameter, PCT the calculated pixel parameter, and P(f) the CDF. Therefore, the pre-processed image appears after contrast enhancement: Iimpre which is further applied to the multilevel thresholding for lung infected region segmentation. The pre-processed images are presented in Fig. 5. As compared to the original CT images, the green channel and CLAHE enhance the image clarity.
3.2 Image segmentation using multilevel thresholding
In this subsection, The mathematical model of the image thresholding algorithm, which uses Otsu’s method is explained [34]. Using image histograms as the input, this method calculates the optimal threshold values for segmenting an image depending on the distribution of pixels across an image. The process provides the best threshold values for CT scan images by increasing the variance between classes to their maximum value, and by computing the optimal threshold values which split the CT images into many groups. The intensity levels of the grey image Gν are identified in the first step of the algorithm and the probability distribution is determined by using Eq. (2) as follows [25].
where il the mentioned intensity level in the range (0 ≤ il ≤ Gv − 1) denotes the total number of pixel values Ni and represents the intensity number il in the histogram of an input image. The histogram is standardized CNi. In Eq. (3), segmentation classes are computed using probability distributions (th) and threshold values.
For A1 and A2, ω0(th) and ω1(th) are additive probability distribution values. These probabilities are defined in Eq. (4) as follows:
The average intensity levels must be defined at this stage as μ0 and μ1 using Eqs. (5) and (6) respectively.
where \( \omega (k)=\sum \limits_{i=1}^k{C}_{N_i} \) and \( \mu (k)=\sum \limits_{i=1}^k{iC}_{N_i} \), and σ1 = ω0(μ0 + μr)2 σ2 = ω1(μ1 + μr)2. Also, μT = ω0μ0 + ω1μ1 and ω0 + ω1 = 1 based on σ1 and σ2.
An optimal set of thresholds are obtained by maximizing the threshold from Eqs. (5) and (6)
Where 1 ≤ th ≤ L − 1, and the Otsu variance, in terms of thresholds, is defined as \( {\sigma}_B^2(th) \).
For multiple thresholds, Eq. (7) can be modified as:
where 1 ≤ thi ≤ L − 1, i = [1, 2, …, p]. The multiple threshold vector is thrs = [th1, th2, …, thp − 1] and L indicates the maximum grey level.
The maximum variance is given by Eq. (8)
For a specific class i, ωi, μj denote the probability of an event’s occurrence and its average. For CT image thresholding at the multilevel these terms are defined as:
3.3 Texture analysis
The distributions and relationships between the image pixels are represented by textures in statistical texture feature extraction algorithms. It ignores the spatial interaction between pixels while computing the average or variance of each pixel. Two or more pixels from different locations may have different properties when they are compared using second-order and higher-order statistics. A textural analysis method derived from GLCMs is the most accepted second-order statistical feature and it is described in detail in the following section.
The second-order statistical features in the form of texture features and co-occurrence matrix were first introduced by Haralick [37]. A 2-step algorithm was introduced with the first step of calculating GLCM and the second step of computing texture features using the calculated GLCMs. For biomedical image analysis, this algorithm has immense potential owing to the quality of its features [8].
Mathematically, the probability of connecting two pixels p and q, with distance d and spatial relation θ is denoted as Pd, θ(p, q). The element (p, q) of P(d, θ) is occurrences of a couple of grey levels p and q with distance d with spatial value θ. A GLCM for an image size N × M with Ng grey levels is a “2D array” of size Ng × Ng. As an example, Fig. 3 shows a GLCM for the size of the image 5 × 4 and Ng = 5. Now, 2D array calculation is using d = 1 and θ= 0. By dividing each element of the co-occurrence matrix by the number of pixels in an image, the matrix is normalized. The GLCMs are defined in all eight directions viz., 0, 45, 90, 135, 180, 225, 270, 315, as shown in Fig. 4. Fine and coarse textures require small and large values d respectively. In MATLAB [45], the operation of the GLCM with 4 orders is shown in Fig. 3. In the current study, a similar type of array structure algorithm is defined for CT image analysis.
To illustrate the GLCM technique used in this work, the computation of the first three values in a GLCM using graycomatrix is as shown in Fig. 2. The values 1 and 1 are assigned to two horizontally adjacent pixels respectively, and the output GLCM contains the value 1 for element (1,1). GLCM comprises the value 1 because the elements (1,1) only appear once in the input CT image. Two neighboring pixels on either side of a horizontal axis have a value of 1, as two adjacent pixels on either side of a vertical axis. A horizontally adjacent pixel with values 1 and 2 in the GLCM (1,2) is represented by the value 2 as there are two separate instances of this. As there are no horizontal adjacent pixels with the values 1 and 3 in the GLCM, element (1,3) is set to 0 because no horizontally adjacent pixels have the value 1. In addition to this, the input image graycomatrix will be processed by scanning for the remaining pixel pairs (p, q), and sums will be recorded in the corresponding GLCM elements.
Figure 3 shows the spatial relationships between the pixels shown by the array of offset values, where ‘D’ stands for the distance from the pixel of interest.
4 Research methodology
COVID-19 patients’ CT images show rapidly spreading patches of opacity accompanied by septal thickening and/or reticulation, a crazy-paving pattern, as well as pleural thickening on an air bronchogram. The lung injury of viral pneumonia is characterized by rapid change in the injury. To classify these changes, a well-patterned feature set is essential in the current work. This section details the extracted feature types and their optimal selection using meta-heuristic algorithms. Further, in this section classification performance of the multi-class LSSVM classifier is discussed for the same image dataset.
4.1 Feature extraction
Once the CT image co-occurrence as the second level histogram is computed, we describe the feature set with 1) LBP and 2) entropy features. LBP was proposed by the authors [48] to reveal local features which are a two-level version. In this method, the size of one pixel on an image is compared with that of the neighboring pixels. The recent modifications in the LBP technique such as selection of neighborhood, enhancement of discriminative capability and robustness, and extension to 3-D data have improved the performance in different applications within the biomedical domain.
The steps for image analysis are deduced using LBP operation to the CT images are shown in Fig. 2. The purpose of using LBP features in this work is to analyze the effect of CT image features as input to LSSVM and CNN when compared to the original images. Additionally, the study increases the depth of image features used in the new algorithm for generating results. In this work, a CT image histogram to label texture descriptors for the LBP codes is used.
In the first step, the labeled image (InABN(x, y)) histogram is used as a descriptor which is defined in Eq. (11),
here I(Im) = 0 then Im is false, I(Im) = 1 is true, and n is total number of distinct labels defined using LBP operator. Here, a circular neighborhood round a pixel with P0 points that are taken on the circumference of a circle having radius Ra. To define the “uniformity metric (U)” as in Eq. (12), \( {LBP}_{P_0R{}_a} \) considered.
Further, a rotation-invariant uniform pattern is obtained with the P0 bits binary number (\( {K}_{P_{0-1}},{K}_{P_{0-2}},\dots, {K}_1,{K}_2 \)) with improved angular quantization. The rotation invariant uniform LBP can be computed as given in Eq. (13)
where gνpp0 denotes the grey value of the center pixel and gνpp0, p0 = 0, . …P0 − 1 indicates the grey values of p0. An LBP image is created for each scale to simplify the size of the texture spectrum histogram. Further, the advantage of the feature descriptor term in terms of energy and the entropy of the LBP image for dissimilar scales corresponding pixel is used in the algorithm.
In biomedical image analysis, a commonly used texture descriptor is the Laws’ feature parameter which represents the image features without considering the frequency domain [11]. One-dimensional vectors with five-pixel lengths are used to obtain the entire mask. In addition, L5 indicates level detection, S5 indicates spots, E5 indicates edges, R5 represents ripples, and W5 indicates waves. When the feature set is to be equipped with several masks of fitting sizes, the Laws’ masks prove to be informative for discriminating between dissimilar types of texture. The method is based on texture energy transforms applied to the image for estimating energy within the pass region of filters. After that, each 2-D mask is applied to the image to extract texture information IimLIR(p, q). While the filter L5E5 is used, the developed texture image is as presented in Eq. (14). All 2-D masks, without L5L5, have zero average depending on the laws. For this reason, we employ the texture image TEXiL5L5 for transforming the remaining textures TEXi(p, q), as shown in Eq. (15)
After performing the fitting convolution of the computed masks, 25 different groupings are generated, and then the results to TEP are sent, which is calculated by moving the nonlinear window mean of absolute values as presented in Eq. (16). In Eq. (17), fourteen rotationally invariant TEP (TR) from combining the 25 energy descriptors are obtained.
The last feature parameter computed in the classification task of the CT images is entropy, which defines the uncertainty connected with randomness in the input dataset. The proposed work employed two kinds of entropy parameters specifically, Kapur’s and Shannon’s entropy [14, 43]. Assume the input segmented image IimLIR(p, q) consists of \( {N}_{l_g} \) distinct grey values. As such, in Eq. (18), the normalized histogram for an RoI of a specific dimension nl × ml is determined. Additionally, Eq. (19) provides the Shannon entropy equation. Compared to the Shannon entropy, Kaur’s entropy Eq. (20) has a higher dynamic range.
The combination of all the features which are extracted is represented as a single feature vector as shown in Eq. (21)
From the feature vector represented in the above equation, the optimal features using the proposed q-MPA algorithm for COVID-19 classification using chest CT images are selected.
4.2 Optimal feature selection using proposed modified quantum-based (Mq-MPA) algorithm
The development process for a simple and efficient metaheuristic optimization method, the Mq-MPA is explained in this section. In medical image classification, generally, optimization techniques have been applied in two domains namely, deterministic, and stochastic. The gradient or non-gradient-based deterministic techniques utilize the gradient information to reach the global point via mathematical programming tools such as linear and non-linear programming [2, 19]. To avoid being stuck in the local optima is to use stochastic optimization-based metaheuristic techniques that use random variables and operators.
To achieve the objective of Mq-MPA the process begins by setting the initial value for a set of N image feature values within the search space that is constrained by lower and upper boundaries. In the next step, the fitness value for each result is computed which determines the most suitable features for the space. The decision of which features to retain and which to discard is taken after this step. In the last step, the finest results are assigned, and the other results can be updated using the quantization of MPA.
MPA works similarly to most metaheuristics in that the initial solution is uniformly distributed over the space of search. Using Eq. (22) as a starting point, MPA proceeds as follows:
in which the lower bound is illustrated by Xmin, Xmax denotes the higher bounds of variables, and K denotes the random number which is uniformly distributed [0, 1].
The elite and prey matrices are two main matrix terms that we define before introducing the MPA technique for feature selection. The elite matrix term depicts the best predator in the process of ‘search and detect’, with prey using the prey’s position information. Equation (23) gives the lite matrix as follows [2]:
Where, \( {\overrightarrow{X}}^I \) denotes the finest predator vector, and to construct the Elite matrix, it is replicated n several times. Further, d denotes the number of dimensions and n gives the number of search agents. As far as important search agents are concerned, both predators and prey must be considered. In contrast to the predator, prey takes on the predator’s role when it seeks its prey. When the best predator can dominate the better predator by the end of every iteration and then Elite is upgraded.
According to the second matrix, named prey, the predators constantly update their positions per matrix position. Specifically, initialization is the process in which the initial prey is created, from which the rightest one (predator) creates the Elite. And hence the prey matrix is as presented in Eq. (24)
where Xp, q represents the pth dimension of qth prey. The elite and prey matrices affect the MPA performance, and the algorithm optimization procedure is divided into three phases depending on the velocity of prey and predator as follows [19, 38]:
-
1.
If the prey operates quicker than the predator then the ratio will be more.
-
2.
Predator and prey move at almost the same speed when they have the same velocity ratio.
-
3.
When a predator outruns a prey, a minimum velocity ratio is likely to occur.
In detail the phases are explained as follows:
-
Phase one: A predator that moves faster than the prey in the form of Brownian motion would update its position in the first scenario. Thus, for maximum repetition, less than one-third of the iteration results in the following Eqs. (25) and (26).
where ⊗ denotes multiplication elementwise, \( {\overrightarrow{R}}_B \) denotes the Brownian motion, and thus forms a vector having arbitrary numbers. The outcome of the prey’s motion is well-defined by \( {\overrightarrow{R}}_B\otimes \overrightarrow{prey_i} \) the term P = 0.5. R indicates a vector of distributed random numbers [0, 1]. For a maximum iteration of ‘1’, this phase occurs in 1/3rd of the iterations when the step size is large for faster exploration capability.
-
Phase two: Exploration and exploitation play equal roles in this step. The scenario above appears, and the exploration tends to become exploitation when predators and preys are of equal velocity. The individuals are therefore divided into two groups, half are processed for exploration, and the rest for exploitation. In this scenario, prey is considered for exploitation and predators for exploration. Also, in this scenario, the Lévy motion is defined for prey and predators to move according to the Brownian pattern. The iterations, in this case, are between one-third and two-thirds of the maximum and are given as,
here \( {\overrightarrow{R}}_L \) denotes Lévy motion that comprises a vector of arbitrary numbers. An example of a simulation of a prey’s motion is provided by \( {\overrightarrow{R}}_L\otimes \overrightarrow{prey_i} \) the Lévy pattern. The possibility of reaching better exploitation is with the majority of the Lévy distribution when the step size is associated with small steps. With the subsequent half of the populace, the following Eqs. (29), (30), and (31) are used [2, 3].
Where CF is a predator’s movement controlling term. Further, RB is multiplied by the Elite matrix to suggest the predator’s travel in the Brownian model, and based on the predator’s travel, the prey position will be updated in this stage.
-
Phase three: When the speed of the predator is larger than that of the prey, this step is used to explore high exploitation capability. Lévy equations work best when the iterations of the position are greater than two-thirds of the maximum number of iterations. In this phase, the following equations are used.
MPA uses a process other than the stages listed above - the fish aggregation devices (FADs) effect [19], which is related to FADs. Equation (34), which presents a mathematical formulation that discusses the impacts of environmental factors on marine predator behavior, illustrates the same.
here FADs = 0.2 is the probability of FADs effect on the optimization method, U is a binary- vector with arrays having an arbitrary number between 0 and 1. For a generated number < 0.2, the array will be ‘0’, else, it represents a value of ‘1’. A random number r is spread between 0 and 1, \( {\overrightarrow{X}}_{\mathrm{min}} \) representing the lower and \( {\overrightarrow{X}}_{\mathrm{max}} \) the upper bound of the sizes. The arbitrary indices of the prey matrix are r1 and r2.
Another term named marine-memory is defined for relating the competence to consider the best places where the predator has been foraging successfully with the memory saving capability in MPA. In addition to updating the prey position and achieving the FADs impact, further the elite matrix is upgraded. A comparison of every solution from the present iteration against its correspondent in the previous iteration is done to obtain a further upgrade. According to this procedure, the quality of the solution improves by following the iterations. Using this method, predators will remember the location of prey-rich areas in the past with successful foraging since they will be able to recall the prey-rich areas from the past.
The MPA technique explained above is affected by being trapped at the local minima even with single-mode, and simple functions in the image classification. The main reason for the drawback is that the velocity vectors presume very small values as the iterations start. A modification based on quantum mechanics has been presented by changing the motion behavior to generate the initial population [1]. In the quantum model of MPA, the state of a particle is depicted by the wave function ψ(r, t) instead of the velocity.
The wave function is defined as in Eq. (35).
The Fourier expansion is defined as in Eqs. (36) and (37).
Equation (38), defines the dynamic performance of a particle as a superposition principle of homochromatic waves on a planar surface.
Energy and momentum can be correlated and the Probability Density Function (PDF) is computed for the global optimal value as in Eq. (39).
where \( {s}_{i,t}^j \) is the global optimal value. The authors [3] have proposed a modification to Eq. (39). With an initial population value of N as shown in Eq. (40).
where Xi indicates the ith solution for r ranging between 0 to 1 as a random number, and the limits of the search space are denoted as Xmax and Xmin for tth the current iteration.
To find the best solution for minimizing the problem, a step involving computing the fitness value of each possible solution has been proposed. The solution update is achieved using Eq. (40) and is modified to Eq. (41).
When compared to the original MPA techniques, the use in which Lévy flight has been used, the Moptimum term results in the lowest fitness value with Pi representing local attractor and pOptimumi as the optimum position obtained by the ith predator to the present time stamp and gOptimumi indicates the optimum position of each predator at every iteration. Moptimum term in the modified algorithm has introduced an average optimum status of the entire crowd, k ∈ [0, 1]. The random values u and θ are uniformly distributed on [0, 1]. The contraction expansion (CE) coefficient β in Eq. (32) is used to control the convergence rate and the value of β descents from 1 to 0.4 for searching for the global optimum. The local search mechanism of the quantum MPA gets the authority in this process as shown in Eq. (44) [40].
where βmax and βmin are the start and end contraction expansion factor values, and itermax indicates the last iteration number.
The random number generators (RNGs) method is affected by the slow convergence and fixing to the local minima. Since biomedical image analysis demands that the image features be selected for optimal correlation with training samples and suggests a modification in the quantum MPA algorithm by using the pseudo-random generator as an alternative to the RNGs. The following modified algorithm equation is used to replace RNGs [10]. Therefore, a modification of q-MPA is proposed by introducing a pseudo-random generation process as defined in Eq. (45).
where l is the initial parameter, with xp initialized as 0. The term is set as l ≥ 8 for the sufficient set size of sl. The speed of the generator is optimized by setting a single iteration in the current work. This step ensures the speed and security of the generator. The algorithm starts by setting the l value, xp with p = 0 the chaotic map {0, 1, 2, …, l ! − 1}. The next step ν0 = p is set and \( {\nu}_{q+1}=\raisebox{1ex}{${\nu}_q$}\!\left/ \!\raisebox{-1ex}{$l!$}\right. \). The second step is repeated until the required number of bits is generated xp + 1.
4.3 Optimized LSSVM-based detection
The authors of [1] propose a step to calculate fitness values for every key so that the finest key with the lowest fitness values can be found. The use of a set of linear equations for training in SVM in the solution of a quadratic optimization problem is the major change between SVM and LS-SVM. The LS-SVM improves the speed of algorithm convergence and is extensively used in classification.
The training data is (xi, yi), i = 1, 2, 3, …, n, xi is the input variable and yi, the output variable in the LS-SVM method. The optimization conditions of the LS-SVM algorithm are:
where w ∈ H - “weight vector” and H is the “higher dimension space”.
The projection of this factor by the nonlinear function φ(x) from the original space R. An optimal hyperplane can be constructed to solve the proposed classification problem.
Equation (49) is updated for the classification technique.
And b ∈ R is the bias, ξ is the regularization factor. This factor is treated as a penalty factor C in SVM and employed in LS-SVM’s confidence interval adjustment. The MPA algorithm is applied as the SVM parameter optimization process to overcome the limitations of setting the penalty factor C and the kernel function C manually. The fitness function is calculated by using the average recognition rate after threefold cross-validation.
The optimization problems are efficiently solved which are described by Eqs. (46) and (47) by transforming the controlled framework into the unrestricted framework by establishing Lagrange multipliers, i = 1, 2, ..n and the “objective function” is obtained as:
According to the optimal solution of MPA conditions [19], one of the most effective kernel functions in LS-SVM is the radial basis function (RBF). As compared to polynomial kernel functions, the RBF kernel involves relatively few parameters. Kernel function selection has been made using RBF. RBF functions were generally chosen as fixed values, including the width (σ) and the regularization (γ) parameters. Nevertheless, the value of γ and σ influences the correctness of the proposed model and its generalization capability. The decision function for the proposed classification method is given by Eq. (49)
An MPA algorithm is used to optimize the model parameters using the 10-fold cross-validation method. The MPA proved to be a more efficient and able technique than other metaheuristic techniques in parameter optimization.
The 10-fold cross-validation is performed as explained below: To assess a classifier’s performance, the average of the ten test accuracies is referred to as the accuracy of the classifier divides the training set as ten subsets of identical size at first; Next, a single subset is taken as the test set and the rest 9 subsets are trained. For illustrating, the approach of considering binary problems per class (Allwein et al., [5]) for a three-class problem has been used (COVID, Non-COVID, and Pneumonia).
5 Results and discussion
In the following section, we present the dataset, and experimental procedure for COVID-19 image classification using CT images. A comparison of LSSVM performance after the feature selection using the metaheuristic approach, and various performance parameters used to assess the algorithms’ performance is also presented in this section.
5.1 Dataset
For the implementation of the proposed system, 20,183 chest CT images are used from the existing database [13, 41]. Of the total images, 4812 CT images are of the COVID-19 negative class with other pulmonary diseases, and 7252 CT images are of the COVID-19 positive class. Figure 4 shows the CT images of COVID-19 for negative, positive, and Pneumonia cases. The dataset to develop a feature selection algorithm for classifying the chest CT images into three classes namely: COVID-19 negative, positive, and pneumonia.
5.2 Experimental procedure
The developed COVID-19 detection methodology in this experimental environment using Intel(R) i5-7200U CPU 2.50GHz, 16.0GB RWindows 10, and MATLAB 2021b is implemented. Here, the experiment was done on 1229 CT images of COVID-19 negative class with other pulmonary diseases, and 1252 are of COVID-19 positive class. Radiologists have identified in these images the regions that have septal thickening, linear opacity, crazy-paving pattern, reticulation, and pleural thickening on-air bronchograms. A batch size of 25 images is considered for evaluating the feature selection, and a maximum of 150 iterations were permitted. The performance of the developed LSSVM-Mq-MPA is compared over CNN, CNN-ResNet50, CNN-VGGNet, and LSSVM-MPA by assessing the performance measures such as “accuracy, sensitivity, specificity, precision, FPR, FNR, NPV, FDR, F1 score, MCC and computation time”.
5.3 Performance parameters
All the extracted features are fed to LS-SVM classifier in the 10-fold cross-validation (CV) approach. In particular, the performances of the classifiers are graded in one of the four different conditions mentioned as follows:
-
True Positive (TRP): the number of images detected as positive COVID-19.
-
True Negative (TRN): the number of images detected as negative COVID-19.
-
False Positive (FAP): the number of positive COVID-19 images detected as negative.
-
False Negative (FAN): the number of negative COVID-19 images detected as positive. Accuracy (ACC) measures the techniques’ ability to differentiate between COVID-19 positive and negative images. The terms sensitivity (SEN), and specificity (SPE) compute the classifier’s capability to accurately determine COVID-19 positive and negative occurrences respectively. These can be defined as:
-
(i)
Accuracy: “ratio of the readings of exactly predicted to the entire readings”. The accuracy formula is as given in Eq. (50),
TRP and TRN represents “true positives, and true negatives”, respectively. Furthermore, FRP and FRN are “false positives and false negatives”, respectively.
-
(ii)
Sensitivity: “the number of true positives, which are exactly predictable” as given in Eq. (51).
-
(iii)
Specificity: “the number of true negatives, which are determined precisely”. It is given in Eq. (52).
-
(iv)
Precision: “the ratio of positive observations that are predicted exactly to the total number of observations that are positively predicted” which is mathematically given in Eq. (53).
-
(v)
False Positive Rate (FPR): calculated as “the ratio of the count of false positive predictions to the entire count of negative predictions”, as given in Eq. (54)
-
(vi)
False Negative Rate (FNR): indicates “the proportion of positives which yield negative test outcomes with the test”. The calculation is indicated in Eq. (55).
-
(vii)
negative predictive value (NPV): indicates the “probability that subjects with a negative screening test truly don’t have the disease”. It is given in Eq. (56).
-
(viii)
false discovery rate (FDR): defines “the number of false positives in all of the rejected hypotheses”. The numerical formula for FDR is given in Eq. (57).
-
(ix)
F1 score: is indicated as the “harmonic mean between precision and recall. It is used as a statistical measure to rate performance”. It is given in Eq. (58)
-
(x)
Matthews Correlation Coefficient (MCC): is indicated as the “correlation coefficient computed by four values”. It is denoted in Eq. (59).
The sample result images of green channel selection, CLAHE, multi-level thresholding, Gray-scale co-occurrence, Gabor filtering, and entropy features of COVID-19, normal, and pneumonia images are shown in Fig. 5.
Three thousand three hundred feature vectors extracted from CT images are used as experimental data. The COVID-19 feature vector is categorized into three groups: COVID-19, normal, and pneumonia. As training samples, the arbitrarily selected 70% sample of each kind of feature vector is used, while the 30% sample of each kind is used as a test sample. Here, the initial parameters of the algorithm are selected following detailed testing. To obtain the most accurate preliminary values of these parameters, Mq-MPA functions were used to alter and select the feature parameters in the simulation. Based on these optimal values of the parameters the final feature set is computed, and the proposed image classification runs at the quickest speed possible.
So, for the classification, the selected feature values of 200 per epoch, maximum iterations of 200, the initial weight of 0.6, and the initial learn factor of 2 are proposed.
A linear adjustment of the learning factor is combined with a quadratic adjustment of the LS-SVM form to produce an improved Mq-MPA algorithm.
Figure 6 displays performance analysis graphs for the classification of normal, pneumonia, and COVID-19 CT images in terms of accuracy, sensitivity, specificity, precision, FPR, FNR, NPV, FDR, F1 score, and MCC using conventional and proposed metaheuristic-based LS-SVMs.
Using the Mq-MPA-based method and the LS-SVMM optimization, Table 2 shows how the performance of the proposed method compares to that of existing classifiers. The accuracy of the recommended LS-SVM-Mq-MPA method is correctly identified in Table 2. Its accuracy is 5.23% less compared to CNN however the time complexity is 22.66% improved than CNN. The proposed modification in the feature space of LS-SVM has also shown reduced computation time by 0.82% when compared to LS-SVM. A 1.39% improved accuracy has been reported by CNN-ResNet50 than LS-SVM-Mq-MPA, and 0.84% improved than CNN-VGGNet. Similarly, the precision of the CNN is 18.18% improved than the LS-SVM-Mq-MPA, 17.05% better in CNN-ResNet50, and 15.45% better than CNN-VGGNet. This implies a better performance for detecting CT abnormalities with the improved LSSVM-Mq-MPA.
The performance of the classifier which used optimized feature space is carried out using Kruskal–Wallis statistical test. Based on the information, the test results are interpreted as follows: the test results based on the measure of uncertainty are in the range 12.27 to 12.32 where the level of significance α =0.05. Therefore, the chance of rejecting the null hypothesis when it is true is 0.05, and the probability of acceptance of the null hypothesis is 0.95 with a probability of uncertainty of 0.002.
Performance comparison of LSSVMs based on conventional and proposed technologies for COVID-19 detection.
The percentage of time-saving per batch plays a very important role when the training and execution time is at most priority. Thereby considering the same scope as the priority, it is observed that there was a significant increase of around 22.66% in the timesaving parameter in the case of the LSSVM-Mq-MPA proposed model to the CNN model. The same can be observed in Fig. 7a where the performance analysis in terms of the percentage of time-saving per batch for the proposed model over the conventional models has been plotted.
In the future, the proposed model can be designed with a collective data model so that the classification of the multidimensional dataset can be addressed with enhanced accuracy. Further, we propose to design a portable embedded handheld device for remote monitoring and diagnosis of COVID-19 in the future.
6 Conclusion
Here, an automated method that detects COVID-19 by assessing CT abnormalities such as septal thickening, linear opacity, crazy-paving pattern, reticulation, and air bronchogram pleural thickening in areas affected by COVID-19 is described. In this work, the processing stages of CT images included pre-processing, septal region removal, air bronchogram pleural region removal, segmentation of abnormalities, and classification. The main contribution of this work is a unique approach to feature selection using the metaheuristic algorithm that provided enhanced diagnostic accuracy for multi-class classification using LSSVM. The modified quantum-based Marine Predators Algorithm improved the performance of COVID-19 image classification accuracy by 12.6% and by 8.73% compared to the CNN model. The ITR, which is crucial in the embedded system development of the system, showed an improvement of 27.4% when compared with a method without feature selection. The work carried out can be extended in the future to segment abnormalities within variants of COVID and to optimize an algorithm to reach high diagnostic rates when classifying the variants. The optimized selection of image features using the proposed algorithm increases the accuracy and automates the process of diagnostics by involving minimum efforts from the radiologists. The accuracy of diagnosis using the proposed method is enhanced when compared to a pure manual method of diagnostics and this encourages work in areas where CT image analysis.
Data availability
The data that support the findings of this study are openly available in repository at https://doi.org/10.1101/2020.04.24.20078584
References
Abd Elaziz M, Mohammadi D, Oliva D, Salimifard K (2021) Quantum marine predators algorithm for addressing multilevel image segmentation. Appl Soft Comput 110:107598. https://doi.org/10.1016/j.asoc.2021.107598
Abd M, Babu S, Anwar I, Lu S, Nastasi B, Alotaibi MA, Hossain A, Yousri D (2021a) Enhanced marine predators algorithm for identifying static and dynamic photovoltaic models parameters. Energy Convers Manag 236(February):113971. https://doi.org/10.1016/j.enconman.2021.113971
Abd M, Mohammadi D, Oliva D, Salimifard K (2021b) Quantum marine predators algorithm for addressing multilevel image segmentation. Appl Soft Comput 110:107598. https://doi.org/10.1016/j.asoc.2021.107598
Ahuja S, Panigrahi BK, Dey N, Rajinikanth V, Gandhi TK (2021) Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl Intell 51(1):571–585. https://doi.org/10.1007/s10489-020-01826-w
Allwein EL, Schapire RE, Singer Y (2000) Reducing multiclass to binary: a unifying approach for margin classifiers. J Mach Learn Res 1:113–141
Arpaci I, Huang S, Al-Emran M, Al-Kabi MN, Peng M (2021) Predicting the COVID-19 infection with fourteen clinical features using machine learning classification algorithms. Multimed Tools Appl 80(8):11943–11957. https://doi.org/10.1007/s11042-020-10340-7
Bahel V, Pillai S (2020) Detection of COVID-19 using chest radiographs with intelligent deployment architecture. In: Big data analytics and artificial intelligence against COVID-19: innovation vision and approach, pp 117–130. https://doi.org/10.1007/978-3-030-55258-9_7
Bannigidad P, Deshpande A (2019) Exudates detection from digital fundus images using GLCM features with decision tree classifier, pp 245–257. https://doi.org/10.1007/978-981-13-9184-2_22
Bhargava A, Bansal A (2021) Novel coronavirus (COVID-19) diagnosis using computer vision and artificial intelligence techniques: a review. Multimed Tools Appl 80(13):19931–19946. https://doi.org/10.1007/s11042-021-10714-5
Caponetto R, Fortuna L, Fazzino S, Xibilia MG (2003) Chaotic sequences to improve the performance of evolutionary algorithms. IEEE Trans Evol Comput 7(3):289–304. https://doi.org/10.1109/TEVC.2003.810069
Chaki J, Dey N (2020) Texture feature extraction techniques for image recognition. Springer Singapore. https://doi.org/10.1007/978-981-15-0853-0
Charte D, Sevillano-García I, Lucena-González MJ, Martín-Rodríguez JL, Charte F, Herrera F (2021) Slicer: feature learning for class separability with least-squares support vector machine loss and COVID-19 chest X-ray case study, pp 305–315. https://doi.org/10.1007/978-3-030-86271-8_26
Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) COVID-19 image data collection: prospective predictions are the future, pp 1–38. http://arxiv.org/abs/2006.11988
Dey N, Rajinikanth V, Fong SJ, Kaiser MS, Mahmud M (2020) Social group optimization–assisted Kapur’s entropy and morphological segmentation for automated detection of COVID-19 infection from computed tomography images. Cogn Comput 12(5):1011–1023. https://doi.org/10.1007/s12559-020-09751-3
Diniz JOB, Quintanilha DBP, Santos Neto AC, da Silva GLF, Ferreira JL, Netto SMB, Araújo JDL, Da Cruz LB, Silva TFB, Caio CM, Ferreira MM, Rego VG, Boaro JMC, Cipriano CLS, Silva AC, de Paiva AC, Junior GB, de Almeida JDS, Nunes RA, … Gattass M (2021) Segmentation and quantification of COVID-19 infections in CT using pulmonary vessels extraction and deep learning. Multimed Tools Appl 80(19):29367–29399. https://doi.org/10.1007/s11042-021-11153-y
Diwan SS, Ravichandran S, Govindarajan R, Narasimha R (2020) Understanding transmission dynamics of COVID-19-type infections by direct numerical simulations of cough/sneeze flows. Trans Indian Natl Acad Eng 5(2):255–261. https://doi.org/10.1007/s41403-020-00106-w
Du R-H, Liang L-R, Yang C-Q et al (2020) Predictors of mortality for patients with COVID19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J 55:2000524. https://doi.org/10.1183/13993003.00524-2020
Elpeltagy M, Sallam H (2021) Automatic prediction of COVID− 19 from chest images using modified ResNet50. Multimed Tools Appl 80(17):26451–26463. https://doi.org/10.1007/s11042-021-10783-6
Faramarzi A, Heidarinejad M, Mirjalili S, Gandomi AH (2020) Marine predators algorithm: a nature-inspired metaheuristic. Exp Syst Appl 152:113377. https://doi.org/10.1016/j.eswa.2020.113377
Gianchandani N, Jaiswal A, Singh D, Kumar V, Kaur M (2020) Rapid COVID-19 diagnosis using ensemble deep transfer learning models from chest radiographic images. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02669-6
Gifani P, Shalbaf A, Vafaeezadeh M (2021) Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int J Comput Assist Radiol Surg 16(1):115–123. https://doi.org/10.1007/s11548-020-02286-w
Hadjinicolaou AV, Farcas GA, Demetriou VL, Mazzulli T, Poutanen SM, Willey BM, Low DE, Butany J, Asa SL, Kain KC, Kostrikis LG (2011) Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch Virol 156(4):671–680. https://doi.org/10.1007/s00705-010-0906-7
Hassanien AE, Mahdy LN, Ezzat KA, Elmousalami HH, Ella HA (2020) Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine. MedRxiv, pp 1–8
Hossam A, Fawzy A, Elnaghi BE, Magdy A (2022) An intelligent model for rapid diagnosis of patients with COVID-19 based on ANFIS. In: Multimedia tools and applications. Springer, pp 338–355. https://doi.org/10.1007/978-3-030-89701-7_30
Houssein EH, Emam MM, Ali AA (2021) Improved manta ray foraging optimization for multi-level thresholding using COVID-19 CT images. Neural Comput & Applic 33:16899–16919. https://doi.org/10.1007/s00521-021-06273-3
Jindal H, Jain S, Suvvari TK, Kutikuppala L, Rackimuthu S, Rocha ICN, Goyal S, Radha (2021) False-negative RT-PCR findings and double mutant variant as factors of an overwhelming second wave of COVID-19 in India: an emerging Global Health disaster. SN Comprehen Clin Med 3(12):2383–2388. https://doi.org/10.1007/s42399-021-01059-z
Kang Z, Li X, Zhou S (2020) Recommendation of low-dose CT in the detection and management of COVID-2019. Eur Radiol 30:4356–4357. https://doi.org/10.1007/s00330-020-06809-6
Kevadiya BD, Machhi J, Herskovitz J et al (2021) Diagnostics for SARS-CoV-2 infections. Nat Mater 20:593–605. https://doi.org/10.1038/s41563-020-00906-z
Kumar H, Fernandez CJ, Kolpattil S, Munavvar M, Pappachan JM (2021 Apr 28) Discrepancies in the clinical and radiological profiles of COVID-19: a case-based discussion and review of literature. World J Radiol 13(4):75–93. https://doi.org/10.4329/wjr.v13.i4.75
Le DN, Subbiah V, Deepak P, Ashish G, Joel K (2021) IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID - 19 diagnosis and classification. Int J Mach Learn Cybern 12(11):3235–3248. https://doi.org/10.1007/s13042-020-01248-7
Loey M, Manogaran G, Khalifa NEM (2020) A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput & Applic 0123456789:1–13. https://doi.org/10.1007/s00521-020-05437-x
Mohanty SK, Satapathy A, Naidu MM et al (2020) Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19) – anatomic pathology perspective on current knowledge. Diagn Pathol, 15:103. https://doi.org/10.1186/s13000-020-01017-8
Nazish, Ullah SI, Salam A, Ullah W, Imad M (2021) COVID-19 lung image classification based on logistic regression and support vector machine. In: Lecture notes in networks and systems: vol 239 LNNS (Issue June), pp 13–23. https://doi.org/10.1007/978-3-030-77246-8_2
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66. https://doi.org/10.1109/TSMC.1979.4310076
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121:103792. https://doi.org/10.1016/j.compbiomed.2020.103792
Pisano ED, Zong S, Hemminger BM, Deluca M, Johnston RE, Muller K, Braeuning MP, Pizer SM (1998) Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J Digit Imaging 11(4):193–200
Shahbahrami A, Pham TA, Bertels K (2012) Parallel implementation of gray level co-occurrence matrices and Haralick texture features on cell architecture. J Supercomput 59(3):1455–1477. https://doi.org/10.1007/s11227-011-0556-x
Shaheen MAM, Yousri D, Fathy A, Hasanien HM, Alkuhayli A, Muyeen SM (n.d.) A novel application of improved marine predators algorithm and particle swarm optimization for solving the ORPD problem
Sharma AK (2020) Novel coronavirus disease (COVID-19). Resonance 25(5):647–668. https://doi.org/10.1007/s12045-020-0981-3
Singh MR, Mahapatra SS (2016) A quantum behaved particle swarm optimization for flexible job shop scheduling. Comput Ind Eng 93:36–44. https://doi.org/10.1016/j.cie.2015.12.004
Soares E, Angelov P, Biaso S, Froes MH, Abe K (2020) SARS-CoV-2 CT-scan dataset : a large dataset of real patients CT scans for SARS-CoV-2 identification, pp 1–8
Sornette D, Mearns E, Schatz M et al (2020) Interpreting, analysing and modelling COVID-19 mortality data. Nonlinear Dyn 101:1751–1776. https://doi.org/10.1007/s11071-020-05966-z
Sparavigna AC (2019) Entropy in image analysis. Entropy 21(5):502. https://doi.org/10.3390/e21050502
Suykens J, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Process Lett 9:293–300. https://doi.org/10.1023/A:1018628609742
The MathWorks, I. (2021) MATLAB and statistics toolbox release 2021b. The MathWorks, Inc., Natick. https://www.mathworks.com/
Weigl JAI, Puppe W, Gröndahl B, Schmitt H-J (2000) Epidemiological investigation of nine respiratory pathogens in hospitalized children in Germany using multiplex reverse-transcriptase polymerase chain reaction. Eur J Clin Microbiol Infect Dis 19(5):336–343. https://doi.org/10.1007/s100960050490
Yasar H, Ceylan M (2021) A novel comparative study for detection of Covid-19 on CT lung images using texture analysis, machine learning, and deep learning methods. Multimed Tools Appl 80(4):5423–5447. https://doi.org/10.1007/s11042-020-09894-3
Zhang G, Huang X, Li SZ, Wang Y, Wu X (2004) Boosting local binary pattern (LBP)-based face recognition, pp 179–186. https://doi.org/10.1007/978-3-540-30548-4_21
Zuiderveld K (1994) Contrast limited adaptive histogram equalization. In: Graphics gems IV. Academic Press Professional, Inc., pp 474–485
Acknowledgments
Authors wish to acknowledge the assistance of radiologists and doctors from KLES Dr. Prabhakar Kore Hospital & Medical Research Centre, Belagavi Karnataka India for their assistance during the manual diagnosis procedure.
Author information
Authors and Affiliations
Contributions
Dattaprasad A. Torse contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Dattaprasad A. Torse, Rajashri Khanai, Krishna Pai, and Sridhar Iyer. The first draft of the manuscript was written by Dattaprasad A. Torse and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Torse, D.A., Khanai, R., Pai, K. et al. Optimal feature selection for COVID-19 detection with CT images enabled by metaheuristic optimization and artificial intelligence. Multimed Tools Appl 82, 41073–41103 (2023). https://doi.org/10.1007/s11042-023-15031-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15031-7