Abstract
Depression is a common cause of increased suicides worldwide, and studies have shown that the number of patients suffering from major depressive disorder (MDD) increased several-fold during the COVID-19 pandemic, highlighting the importance of disease detection and depression management, while increasing the need for effective diagnostic tools. In recent years, machine learning and deep learning methods based on electroencephalography (EEG) have achieved significant results in the field of automatic depression detection. However, most current studies have focused on a small number of EEG signal channels, and experimental data require special processing by professionals. In this study, 128 channels of EEG signals were simply filtered and 24-fold leave-one-out cross-validation experiments were performed using 2DCNN-LSTM classifier, support vector machine, K-nearest neighbor and decision tree. The current results show that the proposed 2DCNN-LSTM model has an average classification accuracy of 95.1% with an AUC of 0.98 for depression detection of 6-second participant EEG signals, and the model is much better than 72.05%, 79.7% and 79.49% for support vector machine, K nearest neighbor and decision tree. In addition, we found that the model achieved a 100% probability of correctly classifying the EEG signals of 300-second participants.
Similar content being viewed by others
1 Introduction
Globally, nearly 13% of children, 46% of adolescents, and 19% of adults are struggling with mental illness each year [63]. Major depressive disorder (MDD) is a worldwide prevalent psychiatric illness characterized by persistent low mood, lack of pleasure and thought inhibition, cognitive impairment, and even strong suicidal ideation [24]. The World Health Organization (WHO) estimates that the total number of people with depression worldwide is approximately 322 million [68]. The current clinical approach to the diagnosis of depression has many obvious drawbacks, including patient denial, poor sensitivity, subjective bias, and inaccuracy [62]. Studies have shown that the number of patients with major depressive disorder (MDD) increased several-fold during the COVID-19 pandemic [9], highlighting the importance of disease detection and management of depression, while increasing the need for effective diagnostic tools.
Electroencephalography (EEG) is a powerful and well-recognized tool for recording brain activity [8]. In recent years, it has been widely used to study and diagnose various neurological disorders, such as depression [30, 36], epilepsy [61], obsessive-compulsive disorder [43], seizure prediction [35, 48, 49, 66], Alzheimer’s disease [19], Creutzfeldt-Jakob disease [40], stroke [3], sleep analysis [54], Parkinson’s disease [58], schizophrenia [4], mood state analysis [65] and brain-computer interfaces (BCI) [39, 59]. The resting EEG signal can effectively avoid the interference signals generated when the brain receives instructions [18]. The use of resting EEG signals for automatic detection of depression is of core research and application value.
Today, artificial intelligence techniques play a central role in almost all advanced systems [22, 26]. Traditional machine learning methods, such as Decision Trees [46], K-nearest Neighbors [50], Support Vector Machine [20] and Random Forest [23], among others, have achieved good results in many fields. With the improvement of computational performance and the increase of data size, Deep Learning is widely used in many fields such as image recognition [44, 57, 60], semantic segmentation [25], target detection [11], emotion recognition [15] and predictive analysis [34]. Convolutional neural networks (CNN) are at the core of the current best architectures for recognizing image and video data, mainly due to their ability to learn and extract feature representations that are robust to partial translation and deformation of the input data [29]. Recurrent Neural Networks (RNN) and Long Short-Term Memory Networks (LSTM) have shown state-of-the-art performance in many applications involving time series dynamics. [69] combined these two types of networks for video classification, and achieved better results.
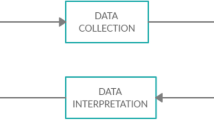
In this study, we propose a hybrid neural network based on EEG signals, i.e., convolutional neural network for temporal learning, windowing and LSTM architecture for sequence learning process for automatic detection of depression on 128-channel resting EEG signals. The general process proposed in this study for preprocessing and classifying 128-channel resting EEG signal data is shown in Fig. 1. The paper makes the following key contributions:
-
We developed a novel hybrid neural network model for automatic detection of depression with 128-channel resting EEG signals.
-
We show that the CNN-LSTM model has better depression detection results compared to the models trained by Decision Trees, K-nearest Neighbors, and Support Vector Machine on the same dataset.
-
We show that the classification of 128-channel EEG signals after simple data processing can reach up to 100% for a single participant, which has not only theoretical but also practical significance for automatic depression detection studies.
The rest of the paper is structured as follows: Section 2 discusses related work associated with this paper; Section 3 describes the dataset used in the study and the data preprocessing process; Section 4 proposes a CNN-LSTM model for depression detection; Section 5 deals with the experiments conducted and is compared and discussed with the results of classical machine learning methods on the same dataset; Section 6 summarizes the conclusions and further research work in the future.
2 Related work
During eye movements (sweeping, blinking, etc.), the electrical field around the eye produces a signal called an Electro-occulogram (EOG). Facial muscle movements produce large amplitude electrical signals called Electromyogram (EMG). EOG and EMG signals are often seen as noise or artifacts in Electro-encephalogram (EEG) signals. [13] reviews some methods for dealing with ocular artifacts in EEG, focusing on the relative merits of various EOG correction procedures. [52] describes the basic concepts of wavelet analysis and other applications. [12] proposed a cascade of three adaptive filters based on the least mean square (LMS) algorithm to reduce common artifacts present in EEG signals without removing the important information embedded in these recordings. The dataset used in this paper is designed to eliminate blink artifacts by using an adaptive noise cancellation technique based on the LMS algorithm.
Machine learning and deep learning methods have shown significant results in the study of automated EEG-based depression detection. Table 1 summarizes relevant studies using EEG signaling for depression screening in recent years. Currently, the traditional machine learning methods for EEG-based depression detection include Decision Tree [27], K-nearest Neighbors [10, 31], Bagged Tree [7], Logistic Regression [21], and Support Vector Machine [5, 33, 38, 42, 45], etc. Deep learning models that have shown excellent results in depression prediction are PNN [2, 17, 37], CNN [1, 32, 41, 53, 67], ANN [16, 47], and CNN-LSTM [6, 51, 56, 64]. [6] proposed a deep hybrid model developed using convolutional neural network (CNN) and long short-term memory (LSTM) architectures, which reported 99.12% and 97.66% classification accuracy for the right and left hemispheres, respectively, for a 2-channel EEG signal. [64] implemented the integration of convolutional neural networks (CNN) and long short-term memory (LSTM) for the classification of depression for 64-channel EEG signals. The accuracy of the left and right hemisphere EEG signals was 99.07% and 98.84%, respectively. [51] applied 1DCNN-LSTM for automatic detection of depression on 19 channels of EEG signals with an accuracy of 99.24%. [56] used a convolutional neural network (CNN) for sequential learning processes with temporal learning, additive windowing, and long short-term memory (LSTM) architecture to screen for depression using 64 channels of EEG signals from 21 depressed and 24 normal subjects, achieving 99.10% accuracy.
In the studies related to EEG-based depression detection, most of them use 2-channel datasets [1, 5, 6, 17, 27, 47, 53], 19-channel datasets [2, 16, 21, 37, 41, 42, 51, 67], and a few studies use 3-channel datasets [10], 8-channel datasets [33, 38], 16-channel datasets [31], 64-channel datasets [56, 64], and 128-channel datasets [32, 45]. [32] proposed a computer-aided detection (CAD) system using a convolutional neural network to study 128 channels of EEG signals from 24 depressed patients and 24 healthy individuals and reported an accuracy rate of 85.62%. [45] collected and analyzed EEG data from 55 subjects at rest, used an altered Kendall rank correlation coefficient and four classification algorithms, and found that the binary linear SVM classifier performed best with a classification accuracy of 92.73% and an AUC of 0.98.
In this study, we performed a 24-fold cross-validation experiment using SVM, K-nearest neighbor, decision tree, and 2DCNN-LSTM classifiers respectively after simple data processing of 128 channels of resting EEG signals from 24 depressed patients and 24 normal participants. The experimental results show that the accuracy of SVM prediction is 72.05%, KNN prediction is 79.7%, decision tree prediction is 79.49%, and CNN-LSTM prediction is 95.1% on the same dataset, which can be seen that CNN-LSTM performs much better than the traditional machine learning methods.
3 Dataset
The EEG signal used in this study was a multimodal open dataset (MODMA Dataset) provided by Lanzhou University for the analysis of mental disorders. Subject data were obtained with the approval of the local biomedical research ethics committee at the Second Hospital of Lanzhou University (Lanzhou, Gansu Province, China). Written informed consent was obtained from all subjects prior to the experiment. Participants included 24 depressed patients (13 males and 11 females; 16–56 years old) and 24 normal subjects (17 males and 7 females; 18–55 years old), and more information about the participants is listed in Table 2.
During the experiment, EEG data were collected from participants with eyes open or closed (resting state) for 5 minutes using a 128-channel HCGSN (HydroCel Geodesic Sensor Net) EEG data acquisition system, and EEG data were recorded using Net Station 4.5.4 software. The sampling frequency was set to 250 Hz for the entire acquisition procedure and the impedance value of the electrodes was below 50 KΩ. The electrodes were placed according to the international standard lead 10–20 electrode system [28] and Cz was set as the reference electrode. 128 channels of electrode placement are shown in Fig. 2. A trap filter was used to suppress the 50 Hz noise, and the EEG signal samples before and after processing are shown in Fig. 3. Blink artifact removal was performed using an adaptive noise cancellation technique based on the LMS algorithm.
After preprocessing the data of all subjects, 75,000 (300 × 250) sampling points from each subject were selected as experimental data according to the sampling frequency of 250 Hz to ensure the consistency of data from all subjects. In this study, the 24-fold leave-one-out cross-validation method was used, and in each fold of the experiment, the training data were 2200 files of 22 depressed and 22 normal subjects, each file containing 1500 sampling points. The validation data and test data were 100 files for 1 depressed and 1 normal subject, respectively, each file containing 1500 sampling points. The experimental procedure of the 24-fold leave-one-out cross-validation method is shown in Fig. 4.
4 Methodology
We took each data file as a subject’s EEG data (6-second resting-state EEG signal of the subject), and then divided each data file into 4-time sequences and inputted them into the model for training analysis, and finally, the model produced a prediction result of “0” or “1” (“0” means the data points belong to non-depressed subjects, while “1” means the data points belong to depressed subjects). The EEG data are segmented with a window length of 1.5 seconds (375 sampling points). Since the sampling rate is 250 samples per second, each EEG segment contains 128 channels and 375 sampling points (window length). the choice of the 1.5-second window size is based on an empirical evaluation of the proposed model, and it is observed that the 1.5-second window size provides the best results.
Figure 5 shows an overview of the 2DCNN-LSTM model we constructed. CNN models are not good at learning sequential information, but are efficient at automatically extracting time-domain features, so we use CNN to extract features from the input EEG signal. The earliest convolutional neural network is the LeNet model proposed by Le Cun [14], and LeNet consists of five main parts, which are the input layer for input images, the convolutional and pooling layers for feature learning, the fully connected layer for integrating the features, and the final output layer of the model. The convolutional layer has three hyperparameters: the size of the convolutional kernel, the step size and the padding. The size of the convolution kernel determines the size of the feature extraction window, the distance that the convolution kernel sweeps sequentially through the input matrix is defined by the step size, and the padding is a way to offset the size shrinkage in the computation. Convolution is calculated as follows:
Where xi denotes the input data, denotes the nth convolution kernel on the ith channel, bn is the bias value, * denotes the convolution operation, and Yn is the nth feature map obtained from the computation.
After each convolution process, the activation function is applied to improve the nonlinearity of CNN. In this paper, the Tanh function is used, and the output range of the Tanh function is known as [−1,1] by Eq. (2), and the output of this activation function is zero mean, which can be used to solve the problem that the output mean of the Sigmoid function is not zero.
Since the traditional recurrent neural network (RNN) is equivalent to a multi-layer feedforward neural network after expansion, the number of layers corresponds to the number of historical data, and too many layers will bring the problem of gradient disappearance (explosion) and loss of historical information during parameter training, so the actual historical information that can be used by the traditional RNN is very limited. In 1997, Hochreiter & Schmidhuber et al. proposed a long short-term memory (LSTM) network to solve the gradient disappearance problem of RNN [55]. The LSTM, as a part of RNN, is usually used for sequence data learning because it not only learns from training, but also remembers what it learned to predict the next element in the sequence as part of the process and feeds the output back to the network. Therefore, the use of LSTM architecture can go for these important features extracted by CNN.
The LSTM network contains three kinds of gates for control - input gate, forget gate, and output gate. The basic unit of the LSTM model is shown in Fig. 6.
-
(1).
forget gate: tanh is selected for the output of the final state, while a function with an output range of [0,1] is selected as the activation function of the gate structure, which determines the degree of retention of the last cell state St − 1. ft value of 0 indicates forgetting and 1 indicates retention, and the calculation formula is as follows:
-
(2).
Input gate: the cell state St at the latest moment is determined by the previous cell state St − 1 and the new cell state to be selected at the moment gt together. ft and it are used as the weight coefficient terms of St − 1and gt, reflecting the update and forgetting of information in the cell, and the calculation equation is as follows:
-
(3).
Output gate: the output gate determines how much of the current state St is output to the current output value ht, calculated as follows:
Table 3 explains in detail each layer of the proposed CNN-LSTM model and the parameters associated with each layer. To train the proposed model, 32 training batch sizes were chosen. The early stopping criterion was used and the best results were saved. Based on the experimental results, we set the convolutional kernel size to 1 × 3 for the convolutional layers, 32 for the first two convolutional layers, and 16 for the last two convolutional layers.
The dropout layer can discard some information of neurons randomly during the training process, which can significantly reduce the overfitting phenomenon. In this model, dropout is set to 0.5, the initial learning rate of the model is 0.01, Adam optimizer is used for optimization, binary cross-entropy is used as the loss function, and the sigmoid activation function is used for binary classification. The role of the sigmoid function is to transform the input into an output between 0 and 1. According to the Eq. (9) of the function, it is known that as x approaches negative infinity, the function approaches to 0, while the value of the function approaches to 1 as x approaches positive infinity. The hyperparameters involved in the CNN-LSTM model proposed in this study are obtained based on the best results of the experiments.
5 Results and discussion
Figure 7 shows the training process of the CNN-LSTM model proposed in this paper in one cross-validation. It can show that with the increase of iterations, the accuracy and AUC of the CNN-LSTM model on both the training and validation sets are gradually increasing, while the loss is gradually decreasing, and the whole training time is very short.
Figure 8 shows the performance of SVM, KNN, decision tree and CNN-LSTM in the 24-fold leave-one-out cross-validation experiments, respectively, and is described in detail in Table 3. According to the results of the experiments, the average accuracy of SVM, KNN, decision tree and CNN-LSTM on the same dataset is 72.05%, 79.7%, 79.49% and 95.1%, respectively, and the highest accuracy is 95.31%, 94.53%, 92.19% and 100%, respectively, and the classification effect of the CNN-LSTM model proposed in this paper is much higher than that of traditional machine learning methods.
The average AUC of the CNN-LSTM model is 0.9803, and the lowest accuracy of 80.21% and the lowest AUC of 0.9044. We further analyzed the experimental results: in each fold of the experiment, the classification of 2 subjects (50 × 2 = 100 test data), the lowest accuracy of classification was 80.21%, i.e., there were at most 20 incorrect predictions during the 24-fold cross-validation, however, each subject actually contained 50 test data, and if we generate a total result for each subject after voting on the 50 predicted results for each subject, then our proposed model will make 100% predictions for each subject.
[32] On the same dataset, a computer-aided detection (CAD) system using convolutional neural network (ConvNet) was proposed and obtained 85.62% accuracy after cross-validating the classification by 24 folds. [45] collected and analyzed 128 channels of resting state EEG data from 55 subjects and used SVM for prediction, obtaining 92.73% accuracy with an AUC of 0.98. The results of the CNN-LSTM model proposed in this paper are as follows: with each subject providing 6 seconds of EEG data, the average prediction accuracy of the model was 95.1% with an AUC of 0.98, the highest prediction accuracy was 100% with an AUC of 1.0, and the lowest prediction accuracy was 80.21% with an AUC of 0.9044. If the subjects provided 300 seconds of EEG data, then after dividing the data into 50 equal parts and inputting them into the model, 50 predictions are obtained, and then voting on the 50 predictions, the final prediction accuracy obtained will be 100% (Table 4).
6 Conclusions
In this study, we propose a 2DCNN-LSTM classifier, and the dataset used for the experiments does not require special processing by professionals, only simple filtering and removal of ocular artifacts for 128 channels of resting EEG signals. The results of the experiments showed that for depression detection of EEG signals of participants with 6 seconds, the average classification accuracy of CNN-LSTM was 95.1% with an AUC of 0.98, and for depression detection of EEG signals of participants with 300 seconds, the classification accuracy of a single participant was 100%. The experimental results outperformed traditional machine learning methods with the same data set and outperformed the literature [32, 45] with similar data sets. In addition, we believe that a model that can perform depression detection without special processing of data will better meet the needs in practical applications, and the research results of this paper have not only theoretical significance but also important practical significance for the study of automatic detection of depression.
As a mainstream research direction in the future, it is necessary to try different network architectures on more datasets to gradually improve our 2DCNN-LSTM model. In the future, we will research other features of EEG signals and other effective data processing and classification methods of depression EEG signals on larger datasets.
Data availability
The datasets analyzed during the current study are available from the MODMA repository, owned by Lanzhou University(http://modma.lzu.edu.cn/data/application//) but restrictions apply to the availability of these data, which were used under license from Lanzhou University, and so are not publicly available. Data are however available from the authors upon reasonable request and permission of the Lanzhou University of Wearable.
References
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H, Subha DP (2018) Automated EEG-based screening of depression using deep convolutional neural network. Comput Methods Prog Biomed 161:103–113
Ahmadlou M, Adeli H, Adeli A (2012) Fractality analysis of frontal brain in major depressive disorder. Int J Psychophysiol 85(2):206–211
Ajčević M, Furlanis G, Naccarato M, Miladinović A, Stella AB, Caruso P et al (2021) Hyper-acute EEG alterations predict functional and morphological outcomes in thrombolysis-treated ischemic stroke: a wireless EEG study. Med Biol Eng Comput 59(1):121–129. https://doi.org/10.1007/s11517-020-02280-z
Akar SA, Kara S, Latifoğlu FATMA, Bilgiç V (2016) Analysis of the complexity measures in the EEG of schizophrenia patients. Int J Neural Syst 26(02):1650008. https://doi.org/10.1142/S0129065716500088
Akbari H, Sadiq MT, Rehman AU (2021) Classification of normal and depressed EEG signals based on centered correntropy of rhythms in empirical wavelet transform domain. Health Inf Sci Syst 9(1):1–15
Ay B, Yildirim O, Talo M, Baloglu UB, Aydin G, Puthankattil SD, Acharya UR (2019) Automated depression detection using deep representation and sequence learning with EEG signals. J Med Syst 43(7):1–12
Bairy GM, Lih OS, Hagiwara Y, Puthankattil SD, Faust O, Niranjan UC, Acharya UR (2017) Automated diagnosis of depression electroencephalograph signals using linear prediction coding and higher order spectra features. J Med Imaging Health Inf 7(8):1857–1862
Binnie CD, Prior PF (1994) Electroencephalography. J Neurol Neurosurg Psychiatry 57(11):1308–1319
Bueno-Notivol J, Gracia-García P, Olaya B, Lasheras I, López-Antón R, Santabárbara J (2021) Prevalence of depression during the COVID-19 outbreak: a meta-analysis of community-based studies. Int J Clin Health Psychol 21(1):100196
Cai H, Han J, Chen Y, et al (2018) A pervasive approach to EEG-based depression detection. Complexity 2018(3):1–13
Chakraborty B, She X, Mukhopadhyay S (2021) A fully spiking hybrid neural network for energy-efficient object detection. IEEE Trans Image Process 30:9014–9029
Correa AG, Laciar E, Patiño HD, Valentinuzzi ME (2007) Artifact removal from EEG signals using adaptive filters in cascade. J Phys Conf Ser 90(1):012081 IOP publishing
Croft R, Barry R (2000) Removal of ocular artifact from the EEG: a review. Neurophys Clin/Clin Neurophys 30(1):5–19
Cun YL (1986) Learning process in an asymmetric threshold network. In disordered systems and biological organization (pp. 233–240). Springer, Berlin, Heidelberg
Do LN, Yang HJ, Nguyen HD, Kim SH, Lee GS, Na IS (2021) Deep neural network-based fusion model for emotion recognition using visual data. J Supercomput 77(10):10773–10790
Erguzel TT, Sayar GH, Tarhan N (2016) Artificial intelligence approach to classify unipolar and bipolar depressive disorders. Neural Comput & Applic 27(6):1607–1616
Faust O, Ang PCA, Puthankattil SD, Joseph PK (2014) Depression diagnosis support system based on EEG signal entropies. J Mech Med Biol 14(03):1450035
Fingelkurts AA, Fingelkurts AA (2015) Altered structure of dynamic electroencephalogram oscillatory pattern in major depression. Biol Psychiatry 77(12):1050–1060
Francis A, Pandian IA (2021) Early detection of Alzheimer’s disease using local binary pattern and convolutional neural network. Multimed Tools Appl 80(19):29585–29600
Guler I, Ubeyli ED (2007) Multiclass support vector machines for EEG-signals classification. IEEE Trans Inf Technol Biomed 11(2):117–126
Hosseinifard B, Moradi MH, Rostami R (2013) Classifying depression patients and normal subjects using machine learning techniques and nonlinear features from EEG signal. Comput Methods Prog Biomed 109(3):339–345
Huh JH, Seo YS (2019) Understanding edge computing: engineering evolution with artificial intelligence. IEEE Access 7:164229–164245
Ikram ST, Priya V, Anbarasu B, Cheng X, Ghalib MR, Shankar A (2022) Prediction of IIoT traffic using a modified whale optimization approach integrated with random forest classifier. J Supercomput 78(8):10725–10756
Jia Z, Huang X, Wu Q, Zhang T, Lui S, Zhang J, Gong Q (2010) High-field magnetic resonance imaging of suicidality in patients with major depressive disorder. Am J Psychiatr 167(11):1381–1390
Kar MK, Nath MK, Neog DR (2021) A review on progress in semantic image segmentation and its application to medical images. SN Comput Sci 2(5):1–30
Khan MA (2017) Multiresolution coding of motion capture data for real-time multimedia applications. Multimed Tools Appl 76(15):16683–16698
Kim AY, Jang EH, Kim S, Choi KW, Jeon HJ, Yu HY, Byun S (2018) Automatic detection of major depressive disorder using electrodermal activity. Sci Rep 8(1):1–9
Klem GH, Lüders HO, Jasper HH, Elger C (1999) The ten-twenty electrode system of the international federation. Int Federation Clin Neurophys Electroencephalogr Clin Neurophysiol Suppl 52:3–6
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Li X, Hu B, Sun S, Cai H (2016) EEG-based mild depressive detection using feature selection methods and classifiers. Comput Methods Prog Biomed 136:151–161
Li Y, Hu B, Zheng X, Li X (2018) EEG-based mild depressive detection using differential evolution. IEEE Access 7:7814–7822
Li X, La R, Wang Y, Niu J, Zeng S, Sun S, Zhu J (2019) EEG-based mild depression recognition using convolutional neural network. Med Biol Eng Comput 57(6):1341–1352
Liao SC, Wu CT, Huang HC, Cheng WT, Liu YH (2017) Major depression detection from EEG signals using kernel eigen-filter-bank common spatial patterns. Sensors 17(6):1385
Liao, W, Zeng, B, Liu, J, Wei, P, Cheng, X (2022) Taxi demand forecasting based on the temporal multimodal information fusion graph neural network. Appl Intell, 1-14
Liu Y, Jiang B, Feng J, Hu J, Zhang H (2021) Classification of EEG signals for epileptic seizures using feature dimension reduction algorithm based on LPP. Multimed Tools Appl 80(20):30261–30282
Loh HW, Ooi CP, Aydemir E, Tuncer T, Dogan S, Acharya UR (2022) Decision support system for major depression detection using spectrogram and convolution neural network with EEG signals. Expert Syst 39(3):e12773
Mahato S, Paul S (2019) Detection of major depressive disorder using linear and non-linear features from EEG signals. Microsyst Technol 25(3):1065–1076
Mantri, S, Patil, D, Agrawal, P, Wadhai, V (2015) Non invasive EEG signal processing framework for real time depression analysis. In 2015 SAI intelligent systems conference (IntelliSys) (pp. 518-521). IEEE
Mohamed EA, Yusoff MZ, Malik AS, Bahloul MR, Adam DM, Adam IK (2018) Comparison of EEG signal decomposition methods in classification of motor-imagery BCI. Multimed Tools Appl 77(16):21305–21327
Morabito FC, Campolo M, Mammone N, Versaci M, Franceschetti S, Tagliavini F, Sofia V, Fatuzzo D, Gambardella A, Labate A, Mumoli L, Tripodi GG, Gasparini S, Cianci V, Sueri C, Ferlazzo E, Aguglia U (2017) Deep learning representation from electroencephalography of early-stage Creutzfeldt-Jakob disease and features for differentiation from rapidly progressive dementia. Int J Neural Syst 27(02):1650039
Mumtaz W, Qayyum A (2019) A deep learning framework for automatic diagnosis of unipolar depression. Int J Med Inform 132:103983
Mumtaz W, Xia L, Ali SSA, Yasin MAM, Hussain M, Malik AS (2017) Electroencephalogram (EEG)-based computer-aided technique to diagnose major depressive disorder (MDD). Biomed Signal Process Control 31:108–115
Özçoban MA, Tan O, Aydin S, Akan A (2018) Decreased global field synchronization of multichannel frontal EEG measurements in obsessive-compulsive disorders. Med Biol Eng Comput 56(2):331–338
Park SW, Huh JH, Kim JC (2020) BEGAN v3: avoiding mode collapse in GANs using variational inference. Electronics 9(4):688
Peng H, Xia C, Wang Z, Zhu J, Zhang X, Sun S, Li J, Huo X, Li X (2019) Multivariate pattern analysis of EEG-based functional connectivity: a study on the identification of depression. IEEE Access 7:92630–92641
Polat K, Güneş S (2007) Classification of epileptiform EEG using a hybrid system based on decision tree classifier and fast Fourier transform. Appl Math Comput 187(2):1017–1026
Puthankattil SD, Joseph PK (2012) Classification of EEG signals in normal and depression conditions by ANN using RWE and signal entropy. J Mech Med Biol 12(04):1240019
Qureshi MB, Afzaal M, Qureshi MS, Fayaz M (2021) Machine learning-based EEG signals classification model for epileptic seizure detection. Multimed Tools Appl 80(12):17849–17877
Ravi, S, Shahina, A, Ilakiyaselvan, N, Khan, AN (2022) Epileptic seizure detection using convolutional neural networks and recurrence plots of EEG signals. Multimed Tools Appl, 1–14
Saeedi M, Saeedi A, Maghsoudi A (2020) Major depressive disorder assessment via enhanced k-nearest neighbor method and EEG signals. Phys Eng Sci Med 43(3):1007–1018
Saeedi A, Saeedi M, Maghsoudi A, Shalbaf A (2021) Major depressive disorder diagnosis based on effective connectivity in EEG signals: a convolutional neural network and long short-term memory approach. Cogn Neurodyn 15(2):239–252
Samar VJ, Bopardikar A, Rao R, Swartz K (1999) Wavelet analysis of neuroelectric waveforms: a conceptual tutorial. Brain Lang 66(1):7–60
Sandheep P, Vineeth S, Poulose M et al (2019) Performance analysis of deep learning CNN in classification of depression EEG signals. In: TENCON 2019-2019 IEEE region 10 conference (TENCON). IEEE, pp 1339–1344
Satapathy, SK, Loganathan, D (2022) Automated classification of multi-class sleep stages classification using polysomnography signals: a nine-layer 1D-convolution neural network approach. Multimed Tools Appl, 1–43
Schmidhuber J, Hochreiter S (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Sharma G, Parashar A, Joshi AM (2021) DepHNN: a novel hybrid neural network for electroencephalogram (EEG)-based screening of depression. Biomed Signal Process Control 66:102393
Sharma N, Gupta S, Mehta P, Cheng X, Shankar A, Singh P, Nayak SR (2022) Offline signature verification using deep neural network with application to computer vision. J Electron Imaging 31(4):041210
Sivaranjini S, Sujatha CM (2020) Deep learning based diagnosis of Parkinson’s disease using convolutional neural network. Multimed Tools Appl 79(21):15467–15479
Sreeja SR, Samanta D (2020) Distance-based weighted sparse representation to classify motor imagery EEG signals for BCI applications. Multimed Tools Appl 79(19):13775–13793
Srinivasan G, Sengupta A, Roy K (2016) Magnetic tunnel junction based long-term short-term stochastic synapse for a spiking neural network with on-chip STDP learning. Sci Rep 6(1):1–13
Sujatha K (2020) Automatic epilepsy detection using hybrid decomposition with multi class support vector method. Multimed Tools Appl 79(15):9871–9890
Sung, M, Marci, C, Pentland, A (2005) Objective physiological and behavioral measures for identifying and tracking depression state in clinically depressed patients. Massachusetts Institute of Technology media laboratory, Cambridge, MA, tech. Rep. TR, 595
Swetaa, A, Gayathri, R, Priya, VV (2019) Awareness of mental health among teenagers. Drug Invent Today, 11(8)
Thoduparambil PP, Dominic A, Varghese SM (2020) EEG-based deep learning model for the automatic detection of clinical depression. Phys Eng Sci Med 43(4):1349–1360
Tonoyan Y, Looney D, Mandic DP, Van Hulle MM (2016) Discriminating multiple emotional states from EEG using a data-adaptive, multiscale information-theoretic approach. Int J Neural Syst 26(02):1650005
Tuncer T, Dogan S, Naik GR, Pławiak P (2021) Epilepsy attacks recognition based on 1D octal pattern, wavelet transform and EEG signals. Multimed Tools Appl 80(16):25197–25218
Uyulan C, Ergüzel TT, Unubol H, Cebi M, Sayar GH, Nezhad Asad M, Tarhan N (2021) Major depressive disorder classification based on different convolutional neural network models: deep learning approach. Clin EEG Neurosci 52(1):38–51
World Health Organization (2017) Depression and other common mental disorders: global health estimates (No. WHO/MSD/MER/2017.2). World Health Organization
Yue-Hei Ng J, Hausknecht M, Vijayanarasimhan S, Vinyals O, Monga R, Toderici G (2015) Beyond short snippets: deep networks for video classification. In proceedings of the IEEE conference on computer vision and pattern recognition, pp 4694–4702
Funding
This research was funded by Cangzhou key research and development plan [204102013].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interests/competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Xu, B. & Yin, H. Depression screening using hybrid neural network. Multimed Tools Appl 82, 26955–26970 (2023). https://doi.org/10.1007/s11042-023-14860-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14860-w