Abstract
A Wireless Body Area Network (WBAN) is a network of wireless sensor nodes that can be installed inside or outside of the human body. This network is crucial in medical problems. Due to storage, battery power, and computational resource limitations, the security of medical information such as medical photographs or other information is a major concern in (WBAN). This work proposes an image encryption approach that addresses these constraints by utilizing adaptive DNA code bases and a new multi chaotic map architecture. DNA coding is used to enhances computing efficiency and gives great data transfer capabilities and the new multi chaotic map was formed by Combining Henon, Gaussian and Logistic map (HGL) To generate more chaotic pseudo-random sequences. Numerous analyses were conducted to test the proposed scheme, such as brute force, statistical, differential assaults and noise added analysis. Results indicated that this scheme has a strong level of security.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The wireless network of the body area is one of most significant emerging health-care technologies. Via it we will keep track of the patient’s wellbeing without affecting his daily life. With regard to WBAN, transmission of patient information must be protected without sacrificing the privacy and confidentiality of the data. Due to any change in data, incorrect diagnosis and treatment can result. The wireless body area network faces many challenges such as memory constraint, computing, usability and scalability [14].
DNA cryptography offers many benefits which make it a promising technology for WBAN [2, 17] First, it has a large parallelism that helps improve the speed of computing. Second, it has wide capacity to store DNA molecules and low energy consumption. So, DNA coding is a modern uninterrupted data technology and provides high protection in WBAN over conventional coding such as DES and AES [11, 16]. And for medical images, several scientists have reported that DNA coding is an effective technique for secure transmission of a medical images [3]. Chaotic encryption technology has characteristics such as vulnerability to initial conditions, pseudo-randomness and non-periodicity, making it commonly used for image encryption [8, 22]. Because of the benefits of DNA coding and the chaotic method, researchers use these two approaches in image cryptosystems to create very effective and stable structures that are difficult to penetrate or crack [4, 6, 20].
1.1 Our involvement
In the proposed scheme adaptive DNA coding is used with a modern one-dimensional (HGL) chaotic mapping system that has more features than the traditional chaotic system (Henon map, Gaussian map, logistic map, …) like larger variable range, wide randomization and several chaotic series. So, the chaotic series generated by (HGL) is hard to prophesy. At first, the medical image input is hashed using SHA-256 hash function to obtain the initial chaotic map (HGL) value and get the chaotic series. At second, the image is crossover and adaptive DNA coding is applied on image and chaotic map to get two DNA matrix. Finally, the two DNA matrix is xored using Xor operation to get a new DNA matrix and DNA decoding rules is implemented to obtain cipher image. The results of simulation and the security analysis show that, the algorithm has been shown to exhibit strong encryption effects and can effectively withstand any type of attack. This article is divided as follow. Section 1 describes the proposed scheme’s preliminaries. Section 2 discusses the suggested scheme for encryption. Section 3 illustrate experimental and simulation tools. Section 4 display security analysis and result. The conclusions are given in Section 5.
2 Preliminaries
2.1 DNA operation
DNA (Deoxyribonucleic acid) is a nucleic acid that holds living organisms’ genetic information. In one DNA sequence, there are four distinct nucleic acid bases: adenine -A, thymine -T, cytosine -C, guanine-G, where complementary pairs are C and G, T and A. We can get 24 encoding schemes if 4 bases A, T, C, G are encoded to 00,11,10,01. Due to the complementary connection between DNA bases, eight types of coding groups comply with the basic complementary rule as shown in Table 1, and there are eight types of XOR operation for each DNA base as well. in Table 2, The XOR operation for DNA rule5 [1].
2.2 Chaotic map
Chaotic maps are extremely sensitive dynamic systems to initial conditions and the parameters of control. These characteristics can be regarded as similar to the required cryptographic structures. The cryptography schemes based on Chaos utilizing boundary conditions as a key to cryptography. The chaotic map as Logistic map, Gaussian map and Henon map are illustrated as the follows, which are simple and standard dynamic equation of complicated chaotic pattern [19, 24].
3 The proposed scheme for encryption
3.1 Overview of proposed algorithm
3.1.1 The suggested multi-chaotic map system (HGL)
The logistic and Gaussian map are merged in this scheme with Henon map as shown in Fig. 1 To obtain a modern one-dimensional chaotic map with a unique design and more variables, parameters and unpredictable attitude. The suggested (HGL) Map’s Mathematical structure is defined in Eq. (1).
Where \({z}_{n}\) ,\({\text{z}}_{\text{n}-1}\)is initial value and a, b, \({\upalpha }\) ,r is the parameter of the chaotic system.
3.1.2 Statistical analysis for (HGL) chaotic system
The randomness of the suggested HGL chaotic map is checked using the NIST test feature consisting of 16 statistical measures. Such measures determine whether or not the sequence generated is random. For such measures, the basic reliance is on the likelihood value (p-value). The value p is compared to the meaning level α which represents the boundary between the region of rejection and non-rejection. The significant level in NIST equals 0.01. It implies that the series is not a random series and deny for p-value less than 0.01 and for p-value more than 0.01, This ensures that the series is random and approved. Table 3 shows the results of the series created by the proposed chaotic map. As a consequence, the suggested sequence of chaotic HST map are totally random and ideal to be used.
3.2 Key generation
For this scheme, the hash function of SHA-256 is applied on the input image to obtain 256-bit of secret key K. In that every single bit alteration in the image would produce a totally different hash value. As in Eq. (2), the secret key K is divided into 8-bit blocks, and the primary values are extracted from Eqs. (3), (4). Since the initial values given are Z0 ' and Z1′.
3.3 Encryption algorithm
-
Step1: image I is hashed to get the initial value of (Henon –Gaussian- Logistic map) according to Section (3.1).
-
Step2: calculate the value of \({\text{H}}_{\text{i}}\)and\({\text{p}}_{\text{i}}\)for plain image by using Eqs. (5), (6). Which is used to determine the DNA rule number used for coding. With it, each image is encoded by a different DNA rule than the other images and then a new image V (m, n) is produced using the crossover technique.
$$ H_i=value\;of\;pixel\;in\;position\;\lbrack1\:\times\:3\rbrack\;+\;value\;of\;pixel\;in\;position\;\lbrack M\;\times \;(N-3)\rbrack$$(5)$$p_i=\;\lbrack Hi\;Mod\;8\rbrack\:+\:1$$(6) -
Step 3: V (m, n) matrix is converted into a binary series by representing each pixel in 8 bits and then coding for the DNA encoding rules according to\({ \text{p}}_{\text{i}}\)for getting\({\text{V}}_{\text{d}\text{n}\text{a}}\)
-
Step 4: create an array of chaotic series W using the suggested chaotic map (HGL) of length (m x n) with parameter and initial value a, b, α, r, Z0, Z1
-
Step 5: Similar to step 2, \({\text{H}}_{\text{c}}\)and \({\text{p}}_{\text{c}}\)for chaotic are calculated and then encode the chaotic array W with DNA encoding rules according to\({\text{p}}_{\text{c}}\) for getting\({ \text{W}}_{\text{d}\text{n}\text{a}}\)
-
Step 6: By using XOR operation in Table 2, \({\text{V}}_{\text{d}\text{n}\text{a}}\)is xor with \({ \text{W}}_{\text{d}\text{n}\text{a}}\) and the outcome is decoding with the selected DNA rule 5 to have the encrypt image Q (m, n)
3.4 Decryption algorithm
-
Step1: The encrypt image is encoded according to the selected DNA rule 5 that was used in the encryption algorithm to obtain \({\text{Q}}_{\text{d}\text{n}\text{a}}\).
-
Step2: Repeat steps 4, 5 as with the encryption algorithm.
-
Step3: Based on the XOR process for DNA in Table 2, \({\text{Q}}_{\text{d}\text{n}\text{a}}\)is xor with\({ \text{W}}_{\text{d}\text{n}\text{a}}\) to get\({ \text{V}}_{\text{d}\text{n}\text{a}}\).
-
Step4: calculate \({\text{p}}_{\text{i}}\)from given\({\text{H}}_{\text{i}}\) according to Eq. (6) and then decoding\({ \text{V}}_{\text{d}\text{n}\text{a}}\) with DNA rule according to\({ \text{p}}_{\text{i}}\) and obtain V.
-
Step5: Inverse crossover to get the plain image I (Figs. 2 and 3).
4 Simulation result and discussion
4.1 Tools
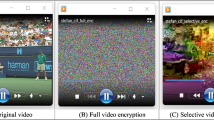
The proposed encryption and decryption scheme are executed on a personal computer using Wolfram (Mathematica11.3) and (MATLAB R2016a) software with Intel ®, core TMi7-8550u, CPU@1.80GH and 8 GB laptop running windows 10. The proposed scheme was conducted on multiple medical images like (MRI, X-Ray, CT, Ultrasound, ECG) images and Lena image with size of 256 × 256 and 512 × 512. Figure 4 display samples of the images tested for encryption and decryption with chaotic control parameter and initial value a = 1.4, b = 0.3, r = 0.399, \({\upalpha }=4.9,\) Z0′= 0.1, Z1′ = 0.15.
4.2 Security analysis and performance tests
4.2.1 Key space analysis
The key space must be wide enough for a successful encryption scheme to stand up to the brute force attack. In this scheme the key is the control parameter (a, b, \({\upalpha }\), r), the initial values (Z0′, Z1′), The value of added pixel of image (\({\text{H}}_{\text{i}}\)) and 256 bits for hash value. If computational precision of number equal 10 − 15. The total key space will be equal 2128 × 10 105 ≈ 3.4028 × 10 143 .This is greater than the permissible space for the key (2256) for realistic symmetrical AES encryption [15]. So, it’s large enough to withstand attack by brute force.
4.2.2 Key sensitivity analysis
Key Sensitivity ensures which only the right key could decode the encrypted image and any effort to decode it for other keys will failure. To evaluate key sensitivity, various key combinations are used to decrypt encrypted images by changing the initial values (Z0′, Z1′) of henon-Gaussian-logistic chaotic map just slightly. As show in Fig. 5, The decryption process by slight change in Z0′and Z1′ does not offer any characteristics of the plain image.
4.2.3 Information entropy analysis
The entropy of the information shall be considered to determine the quantity of randomness in the image. The maximum value of the information entropy for an ideal grayscale image is 8 by Eq. (7). Where m, P (mi) refers to the source of the information and the likelihood of the mi sign. Table 4 demonstrates the entropy results [5].
4.2.4 The histogram analysis
The image histogram shows the spread of pixel value inside the image. against the different intensity levels. The histogram of the encoding image must be uniform enough to be an effective image encoding system. Shows in Fig. 6, the histograms of original and cipher images. The histogram of the images is evidently very different and the histogram of the cipher image is standardized, that allows it to face the statistical assault.
4.2.5 Correlation analysis
This informs how much the relationship is between the similar pixels of the original and the encoded image. Cryptography algorithms should increase the connection between the adjacent pixels in the cipher image. Correlation coefficients for our structure are calculated by Eq. (8) utilizing randomly 4000 pairs of adjacent pixels from both the plain and encrypted image, and the results are shown in Table 6.
The results show that in the original image the correlation coefficients are near to 1while the coefficients of the cipher image are around 0. So that signifies how our scheme has assisted in removing the relationship in the image between neighboring pixels, which enables to avoid statistical attacks as shown in Fig. 6 and Table 5.
4.2.6 Differential attack analysis
The sensitivity of an encrypted image to the few changes in the original image is one way of measuring. The two variables used are NPCR and UACI, which are determined using the Eq. (9). Overall, the NPCR and UACI values should be approximate 99.6093% and 33.4635% respectively. In this scheme one pixel in the plain image was modified and both NPCR and UACI are calculated and from the results showing in Table 6, we notice that NPCR > 99.6% and UACI > 33.3% which make it able to overcome differential attacks [5].
4.2.7 Peak signal to noise analysis
The PSNR is used in the evaluation of image quality, arithmetically described by the mean square error (MSE) as in the Eq. (10). Table 7 shows the value of MSE and PSNR values using the proposed scheme. Results indicate that the PSNR values between both the encoded and the plain images are low and that the MSE values between both the encoded and the plain images are big. Which indicates that this scheme has a strong performance and greater safety.
4.2.8 Noise added analysis
During transmission over a noisy channel, the encrypted image is exposed to noise. The efficiency of the scheme is determined by the noise resistance and the reliable receiver’s capability to identify the image after decoding. By adding Gaussian noise with variance (0.01, 0.1) and pepper and salt noise with density (0.05, 0.5) to the encrypted image, the proposed scheme is checked and then decrypted with appropriate key. Figure 7 displays the encrypted and decoded images.
4.2.9 Complexity
The computational complexity in this scheme shall be determined as the following, for an image with size M x N the time complexity is O(M x N). The algorithm generates a random number with size of M x N using a chaotic map so the complexity for chaotic series is O(M x N), then the complexity time for encoding image and chaotic series to DNA is O(4 x M x N) and the complexity time for DNA XOR operation is O(4 x M x N) also the time of decoding DNA sequence is O(8 x M x N) so the total complexity of the algorithm is nearly equal O(8 x M x N). The computational complexity of the algorithm will also be reduced when the algorithm is run in parallel mode.
4.3 Comparison results with others
The performance of the proposed image coding scheme is compared with other approaches using Entropy, UACI, and NPCR tests and the results demonstrate the fact that our algorithm provides better performance in terms of protection compared to the results obtained by other algorithms.
As shown in Table 8; The entropy of the proposed Lena image is greater than the entropy in the other references and this proves that the proposed algorithm is robust enough and capable of countering statistical attacks.
Table 9 indicates that, relative to the other references, the UACI and NPCR values of the proposed Lena image are good enough and capable of overcoming differential attacks.
Table 10 shows that; Medical images tested with the proposed algorithm have good Entropy, UACI and NPCR values compared to other references making them able to withstand statistical and differential attacks.
5 Conclusion
This paper proposes a medical image encryption scheme using Adaptive DNA and new multi chaotic map (HGL). by using Adaptive DNA each image is encrypted with DNA rule different to other images That makes the algorithm proposed effective against attackers ' perceptions. Whereas a new multi-chaotic map was created by combining Henon, Gaussian and Logistic map (HGL) that produce more chaotic pseudo-random series. the security analysis and experimental Simulation Result show that this algorithm has high entropy value, low correlation, low PSNR value and uniform histogram, high level of security, large key space make it capable of resisting all kinds of attacks.
Change history
27 April 2023
A Correction to this paper has been published: https://doi.org/10.1007/s11042-023-15386-x
References
Abdelfattah RI, Mohamed H, Nasr ME (2020) Secure image encryption scheme based on DNA and new multi chaotic map. J Phys: Conf Ser 1447(1):012053. IOP Publishing
Abo Ajeeb A, Mahmod A, Maala B, Ahmad AS (2017) Enhanced DNA cryptography for wireless body sensor networks. IJIRCCE 5(12)
Akkasaligar PT, Biradar S (2016) Secure medical image encryption based on intensity level using Chao’s theory and DNA cryptography. In: 2016 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), pp 1–6
Akkasaligar PT, Biradar S (2018) Medical image encryption with integrity using DNA and chaotic map. In: International Conference on Recent Trends in Image Processing and Pattern Recognition, Springer, Singapore, pp 143–153
Chen X, Hu CJ (2017) Adaptive medical image encryption algorithm based on multiple chaotic mapping. Saudi J Biol Sci 24(8):1821–1827
Cui G, Wang L, Zhang X, Zhou Z (2018) An image encryption algorithm based on dynamic DNA coding and hyper-chaotic Lorenz system. International Conference on Bio-Inspired Computing: Theories and Applications. 952, pp 226–238
Dagadu JC, Li JP, Aboagye EO (2019) Medical image encryption based on hybrid chaotic DNA diffusion. Wireless personal communications. Springer Sci 108(1):591–612
Dagadu JC, Li J, Aboagye EO, Ge X 2017 Chaotic medical image encryption based on arnold transformation and pseudo randomly enhanced logistic map. J Multidiscip Eng Sci Technol (JMEST) 4:8096–8103
Elamrawy F, Sharkas M, Nasser AM (2018) An image encryption based on DNA coding and 2DLogistic chaotic map. Int J Signal Process 3
Gan G, Lu Y, Chen Y, Han D (2017) A novel image encryption algorithm based on the chaotic system and DNA computing. Int J Mod Phys C. ID28, 1750069
Hossain S, Alam R, Biswas R, Morimoto Y (2016) A dna cryptographic technique based on dynamic DNA sequence table. International Conference on Computer and Information Technology (ICCIT), 51, pp 270–275
Joan D, Vincent R (2002) The design of rijndael AES - the advanced encryption standard. Springer, Berlin Heddberg
Liu Y, Lin T, Wang J, Yuan H (2018) Bit image encryption algorithm based on hyper chaos and dna sequence. J Computers 29:43–55
Marhoon AF, Hamad AH (2015) Chaos theory and DNA computation based data encryption system for E-healthcare monitoring system. Chaos 5(5)
Paul S, Dasgupta P, Naskar PK, Chaudhuri C (2017) Secured image encryption scheme based on DNA encoding and chaotic map. International Information and Engineering Technology Association. 4 pp 70–75
Poriye M, Upadhyaya S (2016) Improved security using dna cryptography in wireless sensor networks. Int J Comput Appl 155:323–325
Raj BB, Vijay JV (2016) Securing data transfer through dna cryptography using symmetric algorithm. Int J Comput Appl 133:19–32
Reyad O, Mofaddel MA, Abd-Elhafiez WM, Fathy M (2017) A novel image encryption scheme based on different block sizes for grayscale and color images. In: 2017 12th International Conference on Computer Engineering and Systems (ICCES). IEEE, pp 455–461
Sahay A, Pradhan C (2017) Multidimensional comparative analysis of image encryption using gauss iterated and logistic maps. In: 2017 International Conference on Communication and Signal Processing (ICCSP). IEEE, pp 1347–1351
Sriramasubramaniam V, Vijayakumar A, Chidambaram N, Thenmozhi K, Rengarajan A, Raj P (2019) A robust 3 tier DNA blended chaotic framework for Grayscale images. In: 2019 International Conference on Computer Communication and Informatics (ICCCI). IEEE, pp 1–6
Sujarani R, Manivannan D (2017) Anon linear two dimensional henon_sine chaotic map based image cryptosystem. Int J Pure Appl Math 115:215–221
Teng L, Wang X, Meng J (2018) A chaotic colour image encryption using integrated bit-level permutation. J Multimed Tools Appl 77:6883–6896
Wang X, Wang Y, Zhu X, Luo C (2020) A novel chaotic algorithm for image encryption utilizing one-time pad based on pixel level and DNA level. Opt Lasers Eng, Elsevier 125:105851
Zhang X, Cao Y (2014) A novel chaotic map and an improved chaos-based image encryption scheme. Sci World J 2014
Zhang X, Han F, Niu Y (2017) Chaotic image encryption algorithm based on bit permutation and dynamic DNA encoding. Computational Intelligence and Neuroscience. ID 63, 6519675, p 11
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The author name "Roayat Ismail Abdefatah" was incorrectly presented as two authors "Roayat Ismail" and "Abdel Fattah" in the original publication of this article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdelfatah, R.I., Saqr, H.M. & Nasr, M.E. An efficient medical image encryption scheme for (WBAN) based on adaptive DNA and modern multi chaotic map. Multimed Tools Appl 82, 22213–22227 (2023). https://doi.org/10.1007/s11042-022-13343-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13343-8