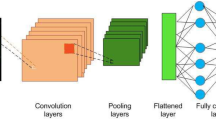
Abstract
Skin Cancer accounts for one-third of all diagnosed cancers worldwide. The prevalence of skin cancers have been rising over the past decades. In recent years, use of dermoscopy has enhanced the diagnostic capability of skin cancer. The accurate diagnosis of skin cancer is challenging for dermatologists as multiple skin cancer types may appear similar in appearance. The dermatologists have an average accuracy of 62% to 80% in skin cancer diagnosis. The research community has been made significant progress in developing automated tools to assist dermatologists in decision making. In this work, we propose an automated computer-aided diagnosis system for multi-class skin (MCS) cancer classification with an exceptionally high accuracy. The proposed method outperformed both expert dermatologists and contemporary deep learning methods for MCS cancer classification. We performed fine-tuning over seven classes of HAM10000 dataset and conducted a comparative study to analyse the performance of five pre-trained convolutional neural networks (CNNs) and four ensemble models. The maximum accuracy of 93.20% for individual model amongst the set of models whereas maximum accuracy of 92.83% for ensemble model is reported in this paper. We propose use of ResNeXt101 for the MCS cancer classification owing to its optimized architecture and ability to gain higher accuracy.







Similar content being viewed by others
References
Abbas Q, Emre Celebi M, Garcia IF, Ahmad W (2013) Melanoma recognition framework based on expert definition of ABCD for dermoscopic images. Skin Research And Technology 19(1):e93–e102. https://doi.org/10.1111/j.1600-0846.2012.00614.x
Alom, MZ, Aspiras, T, Taha, TM, & Asari, VK (2020). Skin cancer segmentation and classification with improved deep convolutional neural network. In: Medical Imaging 2020: Imaging informatics for healthcare, research, and applications, vol. 11318, pp. 1131814. International Society for Optics and Photonics. doi: https://doi.org/10.1117/12.2550146.
Australian Government (2018). Melanoma of the skin statistics. https://melanoma.canceraustralia.gov.au/statistics. Accessed 19 June 2019.
Ballerini L, Fisher RB, Aldridge B, Rees J (2013) A color and texture based hierarchical K-NN approach to the classification of non-melanoma skin lesions. In: Color medical image analysis. Springer, Dordrecht, pp 63–86
Binder M, Schwarz M, Winkler A, Steiner A, Kaider A, Wolff K, Pehamberger H (1995) Epiluminescence microscopy: a useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Arch Dermatol 131(3):286–291
Bishop CM (2006) Pattern recognition and machine learning. Springer
Blum A, Luedtke H, Ellwanger U, Schwabe R, Rassner G, Garbe C (2004) Digital image analysis for diagnosis of cutaneous melanoma. Development of a highly effective computer algorithm based on analysis of 837 melanocytic lesions. Br J Dermatol 151(5):1029–1038. https://doi.org/10.1111/j.1365-2133.2004.06210.x
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Burroni M, Corona R, Dell’Eva G, Sera F, Bono R, Puddu P, Rubegni P (2004) Melanoma computer-aided diagnosis: reliability and feasibility study. Clin Cancer Res 10(6):1881–1886. https://doi.org/10.1158/1078-0432.CCR-03-0039
Cancer Facts and Figures 2016 - American Cancer Society. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2016.html. Accessed 31March 2019.
Celebi ME, Iyatomi H, Stoecker WV, Moss RH, Rabinovitz HS, Argenziano G, Soyer HP (2008) Automatic detection of blue-white veil and related structures in dermoscopy images. Comput Med Imaging Graph 32(8):670–677. https://doi.org/10.1016/j.compmedimag.2008.08.003
Celebi ME, Kingravi HA, Uddin B, Iyatomi H, Aslandogan YA, Stoecker WV, Moss RH (2007) A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph 31(6):362–373. https://doi.org/10.1016/j.compmedimag.2007.01.003
Chaturvedi, SS, Gupta, K, Prasad, P (2019). Skin lesion analyser: an efficient seven-way multi-class skin cancer classification using MobileNet. arXiv preprint arXiv:1907.03220.
Chollet, F. (2015). GitHub - keras-team/keras: Deep Learning for humans. https://github.com/keras-team/keras. Accessed 24 June 2019.
Chollet, F (2017). Xception: deep learning with depthwise separable convolutions. In: IEEE conference on computer vision and pattern recognition, pp. 1251–1258.
Codella N, Cai J, Abedini M, Garnavi R, Halpern A, Smith JR (2015, October) Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In: International workshop on machine learning in medical imaging. Springer, Cham, pp 118–126
Deng, J, Dong, W, Socher, R, Li, LJ, Li, K, Fei-Fei, L (2009). Imagenet: a large-scale hierarchical image database. In: IEEE conference on computer vision and pattern recognition, pp. 248–255. doi: https://doi.org/10.1109/CVPRW.2009.5206848.
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118. https://doi.org/10.1038/nature21056
Fan, DP, Cheng, MM, Liu, JJ, Gao, SH, Hou, Q, Borji, A (2018). Salient objects in clutter: bringing salient object detection to the foreground. In: proceedings of the European conference on computer vision (ECCV), pp. 186-202. Springer, Cham. Doi: https://doi.org/10.1007/978-3-030-01267-0_12.
FAQ - Keras Documentation (2019). https://keras.io/getting-started/faq/#why-is-the-training-loss-much-higher-than-the-testing-loss. Accessed 29 June 2019.
Fu, K, Fan, DP, Ji, GP, Zhao, Q (2020). JL-DCF: joint learning and densely-cooperative fusion framework for RGD-D salient object detection. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 3052–3062.
Fu K, Zhao Q, Gu IYH, Yang J (2019) Deepside: a general deep framework for salient object detection. Neurocomputing 356:69–82
Ge Z, Demyanov S, Chakravorty R, Bowling A, Garnavi R (2017) Skin disease recognition using deep saliency features and multimodal learning of dermoscopy and clinical images. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 250–258. https://doi.org/10.1007/978-3-319-66179-7
Gong, C, Tao, D, Liu, W, Maybank, SJ, Fang, M, Fu, K, Yang, J (2015). Saliency propagation from simple to difficult. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2531–2539.
Goodson AG, Grossman D (2009) Strategies for early melanoma detection: approaches to the patient with nevi. J Am Acad Dermatol 60(5):719–735. https://doi.org/10.1016/j.jaad.2008.10.065
Google Developers (2019). Machine Learning Glossary. https://developers.google.com/machine-learning/glossary. Accessed 24 June 2019.
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32. https://doi.org/10.1016/j.jbi.2018.08.006
Harangi, B, Baran, A, Hajdu, A (2018). Classification of skin lesions using an ensemble of deep neural networks. In: IEEE 40th annual international conference of the IEEE engineering in medicine and biology society - EMBC’2018, pp. 2575–2578. doi: https://doi.org/10.1109/EMBC.2018.8512800.
He, K, Zhang, X, Ren, S, Sun, J (2016). Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition, pp. 770–778.
Iyatomi H, Oka H, Saito M, Miyake A, Kimoto M, Yamagami J, Argenziano G (2006) Quantitative assessment of tumour extraction from dermoscopy images and evaluation of computer-based extraction methods for an automatic melanoma diagnostic system. Melanoma Res 16(2):183–190. https://doi.org/10.1097/01.cmr.0000215041.76553.58
Jana, E, Subban, R, Saraswathi, S (2017). Research on skin Cancer cell detection using image processing. In: IEEE international conference on computational intelligence and computing research - ICCIC’2017, pp. 1–8. doi: https://doi.org/10.1109/ICCIC.2017.8524554.
Kaggle: Your Home for Data Science (2019). https://www.kaggle.com/. Accessed 31 March 2019.
Kasmi R, Mokrani K (2016) Classification of malignant melanoma and benign skin lesions: implementation of automatic ABCD rule. IET Image Process 10(6):448–455. https://doi.org/10.1049/iet-ipr.2015.0385
Kawahara, J, BenTaieb, A, Hamarneh, G (2016). Deep features to classify skin lesions. In: IEEE 13th international symposium on biomedical imaging - ISBI’2016, pp 1397-1400). doi: https://doi.org/10.1109/ISBI.2016.7493528.
Kawahara J, Hamarneh G (2016) Multi-resolution-tract CNN with hybrid pretrained and skin-lesion trained layers. In: International workshop on machine learning in medical imaging. Springer, Cham, pp 164–171. https://doi.org/10.1007/978-3-319-47157-0_20
Kingma, DP, Ba, J (2014). Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980.
Kittler H, Pehamberger H, Wolff K, Binder MJTIO (2002) Diagnostic accuracy of dermoscopy. The lancet oncology 3(3):159–165. https://doi.org/10.1016/S1470-2045(02)00679-4
Koh HK, Geller AC, Miller DR, Grossbart TA, Lew RA (1996) Prevention and early detection strategies for melanoma and skin cancer: current status. Arch Dermatol 132(4):436–443
Korotkov K, Garcia R (2012) Computerized analysis of pigmented skin lesions: a review. Artif Intell Med 56(2):69–90. https://doi.org/10.1016/j.artmed.2012.08.002
Krizhevsky, A, Sutskever, I, Hinton, GE (2012). Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp. 1097–1105.
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Lopez, AR, Giro-i-Nieto, X, Burdick, J, Marques, O (2017). Skin lesion classification from dermoscopic images using deep learning techniques. In: IEEE 13th IASTED international conference on biomedical engineering – BioMed’2017, pp 49-54. doi: https://doi.org/10.2316/P.2017.852-053.
Maglogiannis I, Doukas CN (2009) Overview of advanced computer vision systems for skin lesions characterization. IEEE Trans Inf Technol Biomed 13(5):721–733. https://doi.org/10.1109/TITB.2009.2017529
Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C (2019) Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph 71:19–29. https://doi.org/10.1016/J.COMPMEDIMAG.2018.10.007
Mahbod, A, Schaefer, G, Wang, C, Ecker, R, Ellinge, I (2019). Skin lesion classification using hybrid deep neural networks. In: IEEE international conference on acoustics, speech and signal processing - ICASSP’2019, pp. 1229–1233.
Majtner, T, Bajić, B, Yildirim, S, Hardeberg, JY, Lindblad, J, Sladoje, N (2018). Ensemble of convolutional neural networks for dermoscopic images classification. arXiv preprint arXiv:1808.05071.
Masood A, Ali Al-Jumaily A (2013) Computer aided diagnostic support system for skin cancer: a review of techniques and algorithms. International journal of biomedical imaging 2013:323268–323222. https://doi.org/10.1155/2013/323268
Mhaske, HR, & Phalke, DA (2013). Melanoma skin cancer detection and classification based on supervised and unsupervised learning. In: IEEE international conference on circuits, controls and communications - CCUBE’2013, pp 1-5. doi: https://doi.org/10.1109/CCUBE.2013.6718539.
Milton, MAA (2019). Automated skin lesion classification using ensemble of deep neural networks in ISIC 2018: skin lesion analysis towards melanoma detection challenge. arXiv preprint arXiv:1901.10802.
Morton CA, Mackie RM (1998) Clinical accuracy of the diagnosis of cutaneous malignant melanoma. Br J Dermatol 138(2):283–287
Moura N, Veras R, Aires K, Machado V, Silva R, Araújo F, Claro M (2019) ABCD rule and pre-trained CNNs for melanoma diagnosis. Multimed Tools Appl 78(6):6869–6888. https://doi.org/10.1007/s11042-018-6404-8
Murphy KP (2012) Machine learning: a probabilistic perspective. MIT press
Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, Plewig G (1994) The ABCD rule of dermatoscopy: high prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol 30(4):551–559
Nyíri T, Kiss A (2018) Novel Ensembling methods for dermatological image classification. In: International conference on theory and practice of natural computing. Springer, Cham, pp 438–448
Oliveira RB, Papa JP, Pereira AS, Tavares JMR (2018) Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Comput & Applic 29(3):613–636. https://doi.org/10.1007/s00521-016-2482-6
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/TKDE.2009.191
Parkin DM, Mesher D, Sasieni P (2011) 13. Cancers attributable to solar (ultraviolet) radiation exposure in the UK in 2010. Br J Cancer 105(2):S66–S69. https://doi.org/10.1038/bjc.2011.486
Pathan S, Prabhu KG, Siddalingaswamy PC (2018) Techniques and algorithms for computer aided diagnosis of pigmented skin lesions-a review. Biomedical Signal Processing and Control 39:237–262. https://doi.org/10.1016/j.bspc.2017.07.010
Piccolo D, Ferrari A, Peris KETTY, Daidone R, Ruggeri B, Chimenti S (2002) Dermoscopic diagnosis by a trained clinician vs. a clinician with minimal dermoscopy training vs. computer-aided diagnosis of 341 pigmented skin lesions: a comparative study. Br J Dermatol 147(3):481–486. https://doi.org/10.1046/j.1365-2133.2002.04978.x
Polat K, Koc KO (2020) Detection of skin diseases from Dermoscopy image using the combination of convolutional neural network and one-versus-all. Journal of Artificial Intelligence And Systems 2(1):80–97. https://doi.org/10.33969/ais.2020.21006.
Ramteke NS, Jain SV (2013) ABCD rule based automatic computer-aided skin cancer detection using MATLAB. International Journal of Computer Technology and Applications 4(4):691
Ratul AR, Mozaffari MH, Lee WS, Parimbelli E (2019) Skin Lesions Classification Using Deep Learning Based on Dilated Convolution bioRxiv:860700. https://doi.org/10.1101/860700
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM (2015) Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA dermatology 151(10):1081–1086. https://doi.org/10.1001/jamadermatol.2015.1187
Rosado B, Menzies S, Harbauer A, Pehamberger H, Wolff K, Binder M, Kittler H (2003) Accuracy of computer diagnosis of melanoma: a quantitative meta-analysis. Arch Dermatol 139(3):361–367
Shahin, AH, Kamal, A, Elattar, MA (2018). Deep ensemble learning for skin lesion classification from dermoscopic images. In: IEEE 9th Cairo international biomedical engineering conference - CIBEC’2018, pp 150-153. doi: https://doi.org/10.1109/CIBEC.2018.8641815.
Sharif Razavian, A, Azizpour, H, Sullivan, J, & Carlsson, S (2014). CNN features off-the-shelf: an astounding baseline for recognition. In: IEEE conference on computer vision and pattern recognition workshops, pp. 806–813.
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Silverberg E, Boring CC, Squires TS (1990) Cancer statistics, 1990. CA Cancer J Clin 40(1):9–26
Simonyan, K, Zisserman, A (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
Szegedy, C, Ioffe, S, Vanhoucke, V, & Alemi, AA (2017). Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence, pp. 4278–4284.
Szegedy, C, Vanhoucke, V, Ioffe, S, Shlens, J, Wojna, Z (2016). Rethinking the inception architecture for computer vision. In: IEEE conference on computer vision and pattern recognition, pp. 2818–2826.
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data 5:180161. https://doi.org/10.1038/sdata.2018.161
Vestergaard ME, Macaskill PHPM, Holt PE, Menzies SW (2008) Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol 159(3):669–676. https://doi.org/10.1111/j.1365-2133.2008.08713.x
Wei, J, Wang, S, & Huang, Q (2019). F3Net: fusion, feedback and focus for salient object detection. arXiv preprint arXiv:1911.11445.
White R, Rigel DS, Friedman RJ (1991) Computer applications in the diagnosis and prognosis of malignant melanoma. Dermatol Clin 9(4):695–702
WHO (2017). Skin cancers. https://www.who.int/uv/faq/skincancer/en/index1.html. Accessed 19 June 2019.
Xie, S, Girshick, R, Dollár, P, Tu, Z, & He, K (2017). Aggregated residual transformations for deep neural networks. In: IEEE conference on computer vision and pattern recognition, pp. 1492–1500.
Yu L, Chen H, Dou Q, Qin J, Heng PA (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36(4):994–1004. https://doi.org/10.1109/TMI.2016.2642839
Yu, Z, Ni, D, Chen, S, Qin, J, Li, S, Wang, T, Lei, B (2017). Hybrid dermoscopy image classification framework based on deep convolutional neural network and Fisher vector. In: IEEE 14th international symposium on biomedical imaging - ISBI’2017, pp 301-304. doi: https://doi.org/10.1109/ISBI.2017.7950524.
Zaqout I (2016) Diagnosis of skin lesions based on dermoscopic images using image processing techniques. International Journal Of Signal Processing, Image Processing And Pattern Recognition 9(9):189–204. https://doi.org/10.14257/ijsip.2016.9.9.18.
Zhang M, Qureshi AA, Geller AC, Frazier L, Hunter DJ, Han J (2012) Use of tanning beds and incidence of skin cancer. J Clin Oncol 30(14):1588–1593. https://doi.org/10.1200/JCO.2011.39.3652
Zhao, JX, Liu, JJ, Fan, DP, Cao, Y, Yang, J, Cheng, MM (2019). EGNet: edge guidance network for salient object detection. In: proceedings of the IEEE international conference on computer vision, pp. 8779–8788.
Zoph, B, Vasudevan, V, Shlens, J, Le, QV (2018). Learning transferable architectures for scalable image recognition. In: IEEE conference on computer vision and pattern recognition, pp. 8697–8710.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chaturvedi, S.S., Tembhurne, J.V. & Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed Tools Appl 79, 28477–28498 (2020). https://doi.org/10.1007/s11042-020-09388-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09388-2