Abstract
Background
Salinity is one of the main abiotic factors that restrict plant growth, physiology, and crop productivity is salt stress. About 33% of the total irrigated land suffers from severe salinity because of intensive underground water extraction and irrigation with brackish water. Thus, it is important to understand the genetic mechanism and identify the novel genes involved in salt tolerance for the development of climate-resilient rice cultivars.
Methods and results
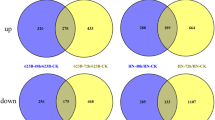
In this study, two rice genotypes with varying tolerance to salt stress were used to investigate the differential expressed genes and molecular pathways to adapt under saline soil by comparative RNA sequencing at 42 days of the seedling stage. Salt-susceptible (S3) and -tolerant (S13) genotypes revealed 3982 and 3463 differentially expressed genes in S3 and S13 genotypes. The up-regulated genes in both genotypes were substantially enriched in different metabolic processes and binding activities. Biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, and plant signal transduction mechanisms were highly enriched. Salt-susceptible and -tolerant genotypes shared the same salt adaptability mechanism with no significant quantitative differences at the transcriptome level. Moreover, bHLH, ERF, NAC, WRKY, and MYB transcription factors were substantially up-regulated under salt stress. 391 out of 1806 identified novel genes involved in signal transduction mechanisms. Expression profiling of six novel genes further validated the findings from RNA-seq data.
Conclusion
These findings suggest that the differentially expressed genes and molecular mechanisms involved in salt stress adaptation are conserved in both salt-susceptible and salt-tolerant rice genotypes. Further molecular characterization of novel genes will help to understand the genetic mechanism underlying salt tolerance in rice.






Similar content being viewed by others
Data availability
The RNA-seq dataset generated and analyzed in the present study is submitted in NCBI GEO under the accession GSE210952.
References
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci 22:123–131
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663
Umali DL (1993) Irrigation-induced salinity: a growing problem for development and the environment. World Bank Publications, Washington, DC
Ahmad H et al (2022) Impact of pre-anthesis drought stress on physiology, yield-related traits, and Drought-responsive genes in green super rice. Front Genet 13:832542
Ali J, Anumalla M, Murugaiyan V, Li Z (2021) Green super rice (GSR) traits: breeding and genetics for multiple biotic and abiotic stress tolerance in rice. In: Ali J, Wani SH (eds) Rice Improvement: physiological, molecular breeding and genetic perspectives. Springer International Publishing, pp 59–97. https://doi.org/10.1007/978-3-030-66530-2_3
Amanat MA et al (2022) Evaluation of green super rice lines for agronomic and physiological traits under salinity stress. Plants 11:1461
Zaid IU et al (2022) Estimation of genetic variances and stability components of yield-related traits of green super rice at multi-environmental conditions in Pakistan. Agronomy 12:1157
Zhang Q (2007) Strategies for developing green super rice. Proc Natl Acad Sci 104:16402–16409
Yu S, Ali J, Zhang C, Li Z, Zhang Q (2020) Genomic breeding of green super rice varieties and their deployment in Asia and Africa. Theor Appl Genet 133:1427–1442
Reddy INBL, Kim B-K, Yoon I-S, Kim K-H, Kwon T-R (2017) Salt tolerance in rice: focus on mechanisms and approaches. Rice Sci 24:123–144
Zhao S et al (2021) Regulation of plant responses to salt stress. Int J Mol Sci 22:4609
Javid MG, Sorooshzadeh A, Moradi F, Modarres SSAM, Allahdadi I (2011) The role of phytohormones in alleviating salt stress in crop plants. Aust J Crop Sci 5:726–734
Zelm EV, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annu Rev Plant Biol 71:403–433
Anwar K, Lakra N, Singla-Pareek SL, Pareek A (2016) Investigating abiotic stress response machinery in plants: the metabolomic approach. In: Dagar JC, Sharma PC, Sharma DK, Singh A (eds) Innovative saline agriculture. Springer, Berlin
Banerjee A, Ghosh P, Roychoudhury A (2019) Salt acclimation differentially regulates the metabolites commonly involved in stress tolerance and aroma synthesis in indica rice cultivars. Plant Growth Regul 88:87–97
Wang X et al (2020) Advances in transcriptomics in the response to stress in plants. Glob Med Genet 7:30–34
Li Y-F et al (2018) Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genomics 19:935
Kong W et al (2019) Meta-analysis of salt stress transcriptome responses in different rice genotypes at the Seedling Stage. Plants 8:64
Wang J et al (2018) Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci Rep 8:2085
Zhou Y et al (2016) Transcriptome analysis of salt stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff). PLoS One 11:e0146242
Ye X, Tie W, Xu J, Ding Z, Hu W (2022) Comparative transcriptional analysis of two contrasting rice genotypes in response to salt stress. Agronomy 12:1163
Chandran AKN et al (2019) Transcriptome analysis of rice-seedling roots under soil–salt stress using RNA-Seq method. Plant Biotechnol Rep 13:567–578
Cartagena JA, Yao Y, Mitsuya S, Tsuge T (2021) Comparative transcriptome analysis of root types in salt tolerant and sensitive rice varieties in response to salinity stress. Physiol Plant 173:1629–1642
Kong W, Sun T, Zhang C, Deng X, Li Y (2021) Comparative transcriptome analysis reveals the mechanisms underlying differences in salt tolerance between Indica and Japonica rice at seedling stage. Front Plant Sci 12:725436
Mirdar Mansuri R et al (2019) Dissecting molecular mechanisms underlying salt tolerance in rice: a comparative transcriptional profiling of the contrasting genotypes. Rice 12:13
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl 30:923–930
Love MI, Huber W, Anders S (2014) Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Pertea M et al (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Drost H-G, Paszkowski J (2017) Biomartr: genomic data retrieval with R. Bioinformatics 33:1216–1217
Carlson, M. & Pagès, H (2021) AnnotationForge: tools for building SQLite-based annotation data packages
Wu T et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation 2:100141
Yanagisawa S (1998) Transcription factors in plants: physiological functions and regulation of expression. J Plant Res 111:363–371
Dobin A et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
McKenna A et al (2010) The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Cingolani P et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6:80–92
Lu Y et al (2020) Coordination between growth-regulating factor1 and GRF-interacting factor1 plays a key role in regulating leaf growth in rice. BMC Plant Biol 20:200
Wang S-L et al (2022) Control of grain weight and size in rice (Oryza sativa L.) by OsPUB3 encoding a U-Box E3 ubiquitin ligase. Rice 15:58
Li W et al (2011) Comparative characterization, expression pattern and function analysis of the 12-oxo-phytodienoic acid reductase gene family in rice. Plant Cell Rep 30:981–995
Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144:1416–1428
Lan T et al (2019) OsSPL10, a SBP-Box Gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L). G3 GenesGenomesGenetics 9:4107–4114
Bhatla SC, Lal MA (2018) Plant physiology development and metabolism. Springer, Berlin
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265
Zhao C, Zhang H, Song C, Zhu J-K, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. The Innovation 1:100017
Formentin E et al (2018) Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00204
Xie Z et al (2020) Characterizing the metabolites related to rice salt tolerance with introgression lines exhibiting contrasting performances in response to saline conditions. Plant Growth Regul 92:157–167
Šamec D, Karalija E, Šola I, Vujčić Bok V, Salopek-Sondi B (2021) The role of polyphenols in abiotic stress response: the influence of molecular structure. Plants 10:118
Shomali A et al (2022) Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants 11:3158
Sharma A et al (2019) Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24:2452
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mata-Pérez C et al (2015) Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00122
Guo J et al (2021) Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int J Mol Sci 22:4921
Abe H et al (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Falcone Ferreyra ML, Rius S, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci 3:222
Qian Y et al (2021) Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front Plant Sci. https://doi.org/10.3389/fpls.2021.677611
Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP-2024R369), King Saud University, Riyadh, Saudi Arabia. We are also thankful to the Bioinformatics and Functional Genomics Labs at the National Institute for Genomics and Advanced Biotechnology (NIGAB), Pakistan, for providing the research facilities.
Funding
No specific funds have been received for this study.
Author information
Authors and Affiliations
Contributions
Conceptualization, NZ, and MRK; methodology, software, and validation, IUZ, UF, MKN, SI, NR, and MU; formal analysis and investigation, NZ, and MU; writing—original draft preparation, review, and editing, NZ, MU, GMA, SF, RMA, JX, ZL, and MRK; visualization, MU; supervision and funding acquisition, MRK. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zahra, N., Uzair, M., Zaid, I.U. et al. The comparative transcriptome analysis of two green super rice genotypes with varying tolerance to salt stress. Mol Biol Rep 51, 22 (2024). https://doi.org/10.1007/s11033-023-08998-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-08998-x