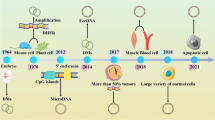
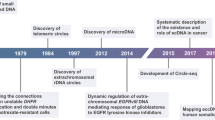
Abstract
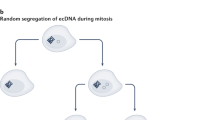
Gene amplification is an increase in the copy number of restricted chromosomal segments that contain gene(s) and frequently results in the over-expression of the corresponding gene(s). Amplification may be found in the form of extrachromosomal circular DNAs (eccDNAs) or as linear repetitive amplicon regions that are integrated into chromosomes, which may form cytogenetically observable homogeneously staining regions or may be scattered throughout the genome. eccDNAs are structurally circular and in terms of their function and content, they can be classified into various subtypes. They play pivotal roles in many physiological and pathological phenomena such as tumor pathogenesis, aging, maintenance of telomere length and ribosomal DNAs (rDNAs), and gain of resistance against chemotherapeutic agents. Amplification of oncogenes is consistently seen in various types of cancers and can be associated with prognostic factors. eccDNAs are known to be originated from chromosomes as a consequence of various cellular events such as repair processes of damaged DNA or DNA replication errors. In this review, we highlighted the role of gene amplification in cancer, the functional aspects of eccDNAs subtypes, the proposed biogenesis mechanisms, and their role in gene or segmental-DNA amplification.




Similar content being viewed by others
References
Albertson DG (2006) Gene amplification in cancer. Trends Genet 22(8):447–455
Matsui A et al (2013) Gene amplification: mechanisms and involvement in cancer. Biomol Concepts 4(6):567–582
Shimizu N (2021) Gene amplification and the extrachromosomal circular DNA. Genes (Basel) 12(10). https://doi.org/10.3390/genes12101533
Wang Y et al (2021) Small ring has big potential: insights into extrachromosomal DNA in cancer. Cancer Cell Int 21(1):236
Verhaak RGW, Bafna V, Mischel PS (2019) Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 19(5):283–288
Paulsen T et al (2018) Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet 34(4):270–278
Wang T et al (2021) Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med 19(1):257
Wang M et al (2021) Extrachromosomal circular DNAs: origin, formation and emerging function in cancer. Int J Biol Sci 17(4):1010–1025
Cao X et al (2021) Extrachromosomal circular DNA: category, biogenesis, recognition, and functions. Front Vet Sci 8:693641
Møller HD et al (2018) Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Natl Commun 9(1):1069
Gaubatz JW (1990) Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res 237(5–6):271–292
Zuo S et al (2021) Extrachromosomal circular DNA (eccDNA): from chaos to function. Front Cell Dev Biol 9:792555
Ling X et al (2021) Small extrachromosomal circular DNA (eccDNA): major functions in evolution and cancer. Mol Cancer 20(1):113
Hotta Y, Bassel A (1965) Molecular size and circularity of DNA in cells of mammals and higher plants. Proc Natl Acad Sci USA 53:356–362
Hamkalo BA et al (1985) Ultrastructural features of minute chromosomes in a methotrexate-resistant mouse 3T3 cell line. Proc Natl Acad Sci USA 82(4):1126–1130
Savelyeva L, Schwab M (2001) Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Lett 167(2):115–123
Shoshani O et al (2021) Chromothripsis drives the evolution of gene amplification in cancer. Nature 591(7848):137–141
Zhu Y et al (2021) Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell 39(5):694–707e7
Wang Y et al (2021) eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature 599(7884):308–314
Oobatake Y, Shimizu N (2020) Double-strand breakage in the extrachromosomal double minutes triggers their aggregation in the nucleus, micronucleation, and morphological transformation. Genes Chromosomes Cancer 59(3):133–143
Yan Y et al (2020) Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance. J Hematol Oncol 13(1):124
Wei J et al (2020) The biogenesis and roles of extrachromosomal oncogene involved in carcinogenesis and evolution. Am J Cancer Res 10(11):3532–3550
Shimizu N, Ochi T, Itonaga K (2001) Replication timing of amplified genetic regions relates to intranuclear localization but not to genetic activity or G/R band. Exp Cell Res 268(2):201–210
Turner KM et al (2017) Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543(7643):122–125
Storlazzi CT et al (2010) Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res 20(9):1198–1206
Storlazzi CT et al (2006) MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet 15(6):933–942
Carroll SM et al (1988) Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol 8(4):1525–1533
Noer JB et al (2022) Extrachromosomal circular DNA in cancer: history, current knowledge, and methods. Trends Genet 38(7):766–781
Schmidt H et al (2009) Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol Rep 22(2):393–400
Cohen S, Regev A, Lavi S (1997) Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene 14(8):977–985
Gu X et al (2020) Novel insights into extrachromosomal DNA: redefining the onco-drivers of tumor progression. J Exp Clin Cancer Res 39(1):215
Cohen Z, Bacharach E, Lavi S (2006) Mouse major satellite DNA is prone to eccDNA formation via DNA ligase IV-dependent pathway. Oncogene 25(33):4515–4524
Basenko EY et al (2010) Telomeric circles are abundant in the stn1-M1 mutant that maintains its telomeres through recombination. Nucleic Acids Res 38(1):182–189
Rivera T et al (2017) A balance between elongation and trimming regulates telomere stability in stem cells. Natl Struct Mol Biol 24(1):30–39
Reddel RR (2003) Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett 194(2):155–162
Zhao S, Wang F, Liu L (2019) Alternative lengthening of telomeres (ALT) in tumors and pluripotent stem cells. Genes (Basel) 10(12). https://doi.org/10.3390/genes10121030
Hull RM, Houseley J (2020) The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet 66(5):889–894
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91(7):1033–1042
Lamming DW (2014) Aging: cellular processes. In: Reference module in biomedical sciences, 3rd edition. Elsevier. https://doi.org/10.1016/B978-0-12-801238-3.00152-5
Haigis MC, Sinclair DA (2011) Sirtuins in aging and age-related diseases Chap. 11. In: Masoro EJ, Austad SN (eds) Handbook of the biology of aging (Seventh Edition). Academic Press, San Diego, pp 243–274
Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13(19):2570–2580
Mehanna P et al (2017) Characterization of the microDNA through the response to chemotherapeutics in lymphoblastoid cell lines. PLoS ONE 12(9):e0184365
Shibata Y et al (2012) Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336(6077):82–86
Bailey C et al (2020) Extrachromosomal DNA-relieving heredity constraints, accelerating tumour evolution. Ann Oncol 31(7):884–893
Paulsen T et al (2019) Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res 47(9):4586–4596
Wu S et al (2019) Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575(7784):699–703
Lin CC et al (1990) Apparent lack of telomere sequences on double minute chromosomes. Cancer Genet Cytogenet 48(2):271–274
Spriggs AI, Boddington MM, Clarke CM (1962) Chromosomes of human cancer cells. Br Med J 2(5317):1431–1435
Lubs HA Jr, Salmon JH (1965) The chromosomal complement of human solid tumors. Ii. karyotypes of glial tumors. J Neurosurg 22:160–168
Cox D, Yuncken C, Spriggs AI (1965) Minute chromatin bodies in malignant tumours of childhood. Lancet 1(7402):55–58
Fan Y et al (2011) Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J Appl Genet 52(1):53–59
Zeng X, Wan M, Wu J (2020) ecDNA within tumors: a new mechanism that drives tumor heterogeneity and drug resistance. Signal Transduct Target Ther 5(1):277
Hanoodi M, Mittal M. Methotrexate (Updated 2023 Jan 16). In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. Available from: https://pubmed.ncbi.nlm.nih.gov/32310574. Accessed 12 July 2022
Meng X et al (2015) Novel role for non-homologous end joining in the formation of double minutes in methotrexate-resistant colon cancer cells. J Med Genet 52(2):135–144
Dillon LW et al (2015) Production of extrachromosomal MicroDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Rep 11(11):1749–1759
Cai M et al (2019) Inhibiting homologous recombination decreases extrachromosomal amplification but has no effect on intrachromosomal amplification in methotrexate-resistant colon cancer cells. Int J Cancer 144(5):1037–1048
Li B, Reddy S, Comai L (2011) Depletion of Ku70/80 reduces the levels of extrachromosomal telomeric circles and inhibits proliferation of ALT cells. Aging 3(4):395–406
Holland AJ, Cleveland DW (2012) Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med 18(11):1630–1638
Shorokhova M, Nikolsky N, Grinchuk T (2021) Chromothripsis-explosion in genetic science. Cells 10(5):1102
Crasta K et al (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482(7383):53–58
Hoffelder DR et al (2004) Resolution of anaphase bridges in cancer cells. Chromosoma 112(8):389–397
Meyerson M, Pellman D (2011) Cancer genomes evolve by pulverizing single chromosomes. Cell 144(1):9–10
Shimizu N et al (1998) Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J Cell Biol 140(6):1307–1320
Von Hoff DD et al (1992) Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc Natl Acad Sci USA 89(17):8165–8169
Vukovic B et al (2007) Correlating breakage-fusion-bridge events with the overall chromosomal instability and in vitro karyotype evolution in prostate cancer. Cytogenet Genome Res 116(1–2):1–11
Tanaka H, Watanabe T (2020) Mechanisms underlying recurrent genomic amplification in human cancers. Trends Cancer 6(6):462–477
Zakov S, Kinsella M, Bafna V (2013) An algorithmic approach for breakage-fusion-bridge detection in tumor genomes. Proc Natl Acad Sci USA 110(14):5546–5551
Tang HL et al (2012) Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell 23(12):2240–2252
Tubio JM, Estivill X (2011) Cancer: when catastrophe strikes a cell. Nature 470(7335):476–477
Ruiz JC et al (1989) Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line. Mol Cell Biol 9(1):109–115
Shimizu N et al (2001) Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res 61(19):6987–6990
Van Roy N et al (2006) Translocation-excision-deletion-amplification mechanism leading to nonsyntenic coamplification of MYC and ATBF1. Genes Chromosomes Cancer 45(2):107–117
Roijer E et al (2002) Translocation, deletion/amplification, and expression of HMGIC and MDM2 in a carcinoma ex pleomorphic adenoma. Am J Pathol 160(2):433–440
Zhang F et al (2009) The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41(7):849–853
Woodcock DM, Cooper IA (1981) Evidence for double replication of chromosomal DNA segments as a general consequence of DNA replication inhibition. Cancer Res 41(6):2483–2490
Osheim YN, Miller OL Jr (1983) Novel amplification and transcriptional activity of chorion genes in Drosophila melanogaster follicle cells. Cell 33(2):543–553
Cohen S, Segal D (2009) Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res 124(3–4):327–338
Goker E et al (1995) Amplification of the dihydrofolate reductase gene is a mechanism of acquired resistance to methotrexate in patients with acute lymphoblastic leukemia and is correlated with p53 gene mutations. Blood 86(2):677–684
Curt GA et al (1983) Unstable methotrexate resistance in human small-cell carcinoma associated with double minute chromosomes. N Engl J Med 308(4):199–202
Sunnerhagen P et al (1986) Molecular cloning and characterization of small polydisperse circular DNA from mouse 3T6 cells. Nucleic Acids Res 14(20):7823–7838
Feng W et al (2022) Targeted removal of mitochondrial DNA from mouse and human extrachromosomal circular DNA with CRISPR-Cas9. Comput Struct Biotechnol J 20:3059–3067
Wang Y, Wang M, Zhang Y (2023) Purification, full-length sequencing and genomic origin mapping of eccDNA. Nat Protoc 18(3):683–699
Acknowledgements
No financial support has been received from any source for this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This study does not require ethical approval. This manuscript does not report any studies, nor any experiments performed on humans or animals by any of the authors.
Informed consent
Not required for this type of manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yüksel, A., Altungöz, O. Gene amplifications and extrachromosomal circular DNAs: function and biogenesis. Mol Biol Rep 50, 7693–7703 (2023). https://doi.org/10.1007/s11033-023-08649-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08649-1