Abstract
Background
Alkaline-salt is one of the abiotic stresses that slows plant growth and developmental processes and threatens crop yield. Long non-coding RNAs (lncRNAs) are endogenous RNA found in plants that engage in a variety of cellular functions and stress responses.
Method
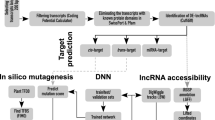
lncRNAs act as competing endogenous RNAs (ceRNA) and constitute a new set of gene control. The precise regulatory mechanism by which lncRNAs function as ceRNAs in response to alkaline-salt stress remains unclear. We identified alkaline-salt responsive lncRNAs using transcriptome-wide analysis of two varieties including alkaline-salt tolerant [WD20342 (WD)] and alkaline-salt sensitive [Caidao (CD)] rice cultivar under control and alkaline-salt stress treated [WD20342 (WDT, and Caidao (CDT)] conditions.
Results
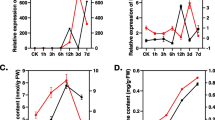
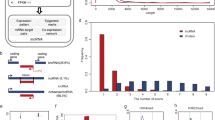
Investigating the competitive relationships between mRNAs and lncRNAs, we next built a ceRNA network involving lncRNAs based on the ceRNA hypothesis. Expression profiles revealed that a total of 65, 34, and 1549 differentially expressed (DE) lncRNAs, miRNAs, and mRNAs were identified in alkaline-salt tolerant WD (Control) vs. WDT (Treated). Similarly, 75 DE-lncRNAs, 34 DE-miRNAs, and 1725 DE-mRNAs (including up-regulated and down-regulated) were identified in alkaline-salt sensitive CD (Control) vs. CDT (Treated), respectively. An alkaline-salt stress ceRNA network discovered 321 lncRNA-miRNA-mRNA triplets in CD and CDT, with 32 lncRNAs, 121 miRNAs, and 111 mRNAs. Likewise, 217 lncRNA-miRNA-mRNA triplets in WD and WDT revealed the NONOSAT000455-osa_miR5809b-LOC_Os11g01210 triplet with the highest degree as a hub node with the most significant positive correlation in alkaline-salt stress response.
Conclusion
The results of our investigation indicate that osa-miR5809b is dysregulated and plays a part in regulating the defense response of rice against alkaline-salt stress. Our study highlights the regulatory functions of lncRNAs acting as ceRNAs in the mechanisms underlying alkaline-salt resistance in rice.






Similar content being viewed by others
Data Availability
All the relevant data are within the paper.
References
Zheng C, Liu C, Liu L et al (2023) Effect of salinity stress on rice yield and grain quality: a meta-analysis. Eur J Agron 144:126765
Islam MA, de Bruyn LL, Warwick NW et al (2021) Salinity-affected threshold yield loss: a signal of adaptation tipping points for salinity management of dry season rice cultivation in the coastal areas of Bangladesh. J Environ Manage 288:112413
Yu Y, Zhou YF, Feng YZ et al (2020) Transcriptional landscape of pathogen-responsive lnc RNA s in rice unveils the role of ALEX 1 in jasmonate pathway and disease resistance. Plant Biotechnol J 18(3):679–690
Wang Y, Luo X, Sun F et al (2018) Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat Commun 9(1):3516
Lu Z, Xia X, Jiang B et al (2017) Identification and characterization of novel lncRNAs in Arabidopsis thaliana. Biochem Biophys Res Commun 488(2):348–354
Severing E, Faino L, Jamge S et al (2018) Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Biol 18(1):1–10
Lu Q, Guo F, Xu Q et al (2020) LncRNA improves cold resistance of winter wheat by interacting with miR398. Funct Plant Biol 47(6):544–557
Li L, Eichten SR, Shimizu R et al (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15:1–15
Zhu M, Zhang M, Xing L et al (2017) Transcriptomic analysis of long non-coding RNAs and coding genes uncovers a complex regulatory network that is involved in maize seed development. Genes 8(10):274
Weidong Q, Hongping C, Zuozhen Y et al (2020) Systematic characterization of long non-coding RNAs and their responses to drought stress in Dongxiang wild rice. Rice Sci 27(1):21–31
Huang X, Zhang H, Wang Q et al (2021) Genome-wide identification and characterization of long non-coding RNAs involved in flag leaf senescence of rice. Plant Mol Biol 105:655–684
Pecrix Y, Staton SE, Sallet E et al (2018) Whole-genome landscape of Medicago truncatula symbiotic genes. Nat Plants 4(12):1017–1025
Gao L, Zhao Y, Ma X et al (2021) Integrated analysis of lncRNA–miRNA–mRNA ceRNA network and the potential prognosis indicators in sarcomas. BMC Med Genom 14:1–11
Meng X, Zhang P, Chen Q et al (2018) Identification and characterization of ncRNA-associated ceRNA networks in Arabidopsis leaf development. BMC Genomics 19:1–10
Wang L, Zhao J, Zhu C et al (2022) Construction of a ceRNA network and comprehensive analysis of lncRNA in hepatocellular carcinoma. Genes 13(5):785
Meng X, Li A, Yu B et al (2021) Interplay between miRNAs and lncRNAs: Mode of action and biological roles in plant development and stress adaptation. Comput Struct Biotechnol J 19:2567–2574
Shin S-Y, Jeong JS, Lim JY et al (2018) Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics 19:1–20
Sun X, Zheng H, Li J et al (2020) Comparative transcriptome analysis reveals new lncRNAs responding to salt stress in sweet sorghum. Front Bioeng Biotechnol 8:331
Zhou R, Sanz-Jimenez P, Zhu X-T et al (2021) Analysis of rice transcriptome reveals the lncRNA/circRNA regulation in tissue development. Rice 14(1):1–16
Franco-Zorrilla JM, Valli A, Todesco M et al (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39(8):1033–1037
Li S, Yu X, Lei N et al (2017) Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep 7(1):45981
Jain P, Hussian S, Nishad J et al (2021) Identification and functional prediction of long non-coding RNAs of rice (Oryza sativa L.) at reproductive stage under salinity stress. Mol Biol Rep 48:2261–2271
Deng F, Zhang X, Wang W et al (2018) Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol 18:1–14
Jiang N, Cui J, Shi Y et al (2019) Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic Res. 6
Li N, Liu H, Sun J et al (2018) Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci Rep 8(1):1–16
Yin M, Ma H, Wang M et al (2021) Transcriptome analysis of flowering regulation by sowing date in Japonica Rice (Oryza sativa L). Sci Rep 11(1):1–11
Bredeson JV, Lyons JB, Oniyinde IO et al (2022) Chromosome evolution and the genetic basis of agronomically important traits in greater yam. Nat Commun 13(1):2001
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom
Wang L, Park HJ, Dasari S et al (2013) CPAT: coding-potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41(6):e74–e
Sun L, Luo H, Bu D et al (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41(17):e166–e
Kong L, Zhang Y, Ye Z-Q et al (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35(suppl2):W345–W9
Chen J, Zhong Y, Qi X (2021) LncRNA TCONS_00021861 is functionally associated with drought tolerance in rice (Oryza sativa L.) via competing endogenous RNA regulation. BMC Plant Biol 21(1):1–12
Yang Z, Yang C, Wang Z et al (2019) LncRNA expression profile and ceRNA analysis in tomato during flowering. PLoS ONE 14(1):e0210650
Ashraf MA, Feng X, Hu X et al (2022) In silico identification of sugarcane (Saccharum officinarum L.) genome encoded microRNAs targeting sugarcane bacilliform virus. PLoS ONE 17(1):e0261807
Wu X, Shi T, Iqbal S et al (2019) Genome-wide discovery and characterization of flower development related long non-coding RNAs in Prunus mume. BMC Plant Biol 19(1):1–17
Uzair M, Long H, Zafar SA et al (2021) Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol 186(1):497–518
Isayenkov SV, Maathuis FJ (2019) Plant salinity stress: many unanswered questions remain. Front Plant Sci 10:80
Hui-Hui Z, Xiu-li Z, Xin L et al (2012) Effects of NaCl and Na2CO3 stresses on the growth and photosynthesis characteristics of Morus alba seedlings. Yingyong Shengtai Xuebao 23(3)
Ferreira TH, Gentile A, Vilela RD et al (2012) microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.).
Feng SJ, Zhang XD, Liu XS et al (2016) Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus. RSC Adv 6(85):82157–82173
Cui J, Jiang N, Hou X et al (2020) Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology 110(2):456–464
Wang A, Hu J, Gao C et al (2019) Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of chinese cabbage (Brassica rapa ssp. chinensis). Sci Rep 9(1):1–14
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP-2023R369), King Saud University, Riyadh, Saudi Arabia. We are thankful to Bioinformatics and Functional Genomics Labs at National Institute for Genomics and Advanced Biotechnology (NIGAB), Pakistan, for providing the research facilities.
Funding
This research was conducted with the funds of the Sino-Pak Agricultural Breeding Innovations Project for Rapid Yield Enhancement (Project #760). The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author information
Authors and Affiliations
Contributions
Conceptualization, O.R., and M.U.; methodology, software, and validation, O.R., B.S., S.F., U.F., and M.U.; formal analysis and investigation, O.R., and M.U.; writing—original draft preparation, review, and editing, O.R., M.S.F., M.U., S.A., S.F., W.H.E.K., I.K., K.A.A., and M.R.K.; visualization, M.U.; supervision and funding acquisition, M.R.K. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Obaid Ur Rehman and Muhammad Uzair equally contributed.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rehman, O.U., Uzair, M., Farooq, M.S. et al. Comprehensive insights into the regulatory mechanisms of lncRNA in alkaline-salt stress tolerance in rice. Mol Biol Rep 50, 7381–7392 (2023). https://doi.org/10.1007/s11033-023-08648-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08648-2