Abstract
Background
Maintaining genetic diversity is of the most essential principle for a long-term conservation of plant genetic resources and could play a crucial role in their management. The genus Aegilops is one important member of wheat germplasm, and there are evidences that novel genes of this genus’ species can be studied/utilized as ideal sources for the wheat cultivar improvement. The objective of this study was to dissect the genetic diversity and population structure among a set of Iranian Aegilops using two gene-based molecular markers.
Methods and results
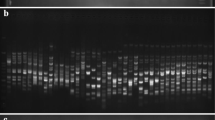
This study investigated the level of genetic diversity among 157 Aegilops accessions consisting of Ae. tauschii Coss. (DD genome), Ae. crassa Boiss. (DDMM genome), and Ae. cylindrica Host. (CCDD genome) belonging to NPGBI using two sets of CBDP and SCoT markers. The SCoT and CBDP primers yielded 171 and 174 fragments, out of which 145 (90.23%) and 167 (97.66%) fragments were polymorphic, respectively. The average of polymorphism information content (PIC)/ marker index (MI)/resolving power (Rp) for SCoT and CBDP markers were 0.32/3.59/16.03 and 0.29/3.01/16.26, respectively. Results of AMOVA revealed the genetic variability within species was greater than the variation observed among them (SCoT: 88% vs. 12%; CBDP: 72% vs. 28%; SCoT + CBDP: 80% vs. 20%). Based on the information obtained from both markers, the higher level of genetic diversity was found in Ae. tauschii as compared to other species. The grouping patterns obtained by Neighbor-joining algorithms, principal coordinate analysis (PCoA), and Bayesian-model-based structure were consistent with each other and resulted in grouping all studied accessions according to their genomic constitutions.
Conclusion
The results of this study revealed a high level of genetic diversity among Iranian Aegilops germplasm. Moreover, SCoT and CBDP marker systems were efficient in deciphering DNA polymorphism and classification of Aegilops germplasm.





Similar content being viewed by others
Data availability
The data in this manuscript are available from the corresponding author upon reasonable request.
Abbreviations
- CBDP:
-
CAAT box-derived polymorphism
- SCoT:
-
Start codon target polymorphism
- NAF:
-
The number of amplified fragments
- NPF:
-
The number of polymorphic fragments
- PIC:
-
Polymorphism information content
- Rp:
-
Resolving power
- MI:
-
Marker index
- PCR:
-
Polymerase chain reaction
- PPL:
-
Percentage polymorphic loci
- Na:
-
The numbers of observed alleles
- Ne:
-
The numbers of effective alleles
- I:
-
Shannon’s information content
- He:
-
Nei’s gene diversity
References
Singh N, Wu S, Tiwari V, Sehgal S, Raupp J, Wilson D, Abbasov M, Gill B, Poland J (2019) Genomic analysis confirms population structure and identifies inter-lineage hybrids in Aegilops tauschii. Front Plant Sci 10:9. https://doi.org/10.3389/fpls.2019.00009
Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT et al (2010) Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11:702. https://doi.org/10.1186/1471-2164-11-702
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. https://doi.org/10.1371/journal.pone.0066428
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 11:1205. https://doi.org/10.3390/plants11091205
Pour-Aboughadareh A, Kianersi F, Poczai P, Moradkhani H (2021) Potential of wild relatives of wheat: ideal genetic resources for future breeding programs. Agronomy 11:1656. https://doi.org/10.3390/agronomy11081656
Nevo E, Chen G (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ 33:670–685. https://doi.org/10.1111/j.1365-3040.2009.02107.x
Khan MK, Pandey A, Hamurcu M, Avsaroglu ZZ, Ozbek M, Omay AH, Elbasan F, Omay MR, Gokmen F, Topal A, Gezgin S (2021) Variability in physiological traits reveals boron toxicity tolerance in Aegilops species. Front Plant Sci 12:736614. https://doi.org/10.3389/fpls.2021.736614
Khan MK, Pandey A, Hamurcu M, Germ M, Yilmaz FG, Ozbek M, Avsaroglu ZZ, Topal A, Gezgin S (2022) Nutrient homeostasis of Aegilops accessions differing in B tolerance level under boron toxic growth conditions. Biology11(8):1094. https://doi.org/10.3390/biology11081094
Kishii M (2019) An update of recent use of aegilops species in wheat breeding. Front Plant Sci 10:585. https://doi.org/10.3389/fpls.2019.00585
Bell G (1987) The history of wheat cultivation in wheat breeding. Springer, Berlin, pp 31–49
Nakai Y (1979) Isozyme variations in Aegilops and Triticum, IV. The origin of the common wheats revealed from the study on esterase isozymes in synthesized hexaploid wheats. Jpn J Genet 54:175–189. https://doi.org/10.1266/JJG.46.321
Hedge SG, Valkoun J, Waines JG (2002) Genetic diversity inwild and weedy Aegilops, Amblyopyrum and Secale species preliminary survey. Crop Sci 42:608–614
Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Ozkan H (2011) Aegilops, Wild Crop relatives, genomic and breeding resources. Cereals, pp 1–76
Hammer K (1980b) Zur Taxonomie und Nomenklatur der Gattung Aegilops L. Feddes Rep 91:225–258. https://doi.org/10.1002/fedr.19800910404
Kam-Morgan L, Gill B, Muthukrishnan S (1989) DNA restriction fragment length polymorphisms: a strategy for genetic mapping of D genome of wheat. Genome 32:724–732. https://doi.org/10.1139/g89-503
Tiwari G, Singh R, Singh N, Choudhury DR, Paliwal R, Kumar A, Gupta V (2016) Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in Kalmegh [Andrographis paniculata (burm. f.) nees]. Ind Crop Prod 86:1–11. https://doi.org/10.1016/j.indcrop.2016.03.031
Puneeth PV, Lata S, Yadav RK, Wankhede DP, Tomar BS, Choudary H, Tomer A, Bidaramali V, Talukdar A (2022) Exploring the genetic diversity using CAAT box-derived polymorphism (CBDP) and start codon targeted (SCoT) markers in cultivated and wild species of okra (Abelmoschus esculentus (L.) Moench). https://doi.org/10.1007/s10722-022-01458-8. Genet Resour Crop Evol
Mostaavi AS, Omidi M, Azizinezhad R, Etminan A, Badi HN (2021) Genetic diversity analysis in a mini core collection of Damask rose (Rosa damascena Mill.) Germplasm from Iran using URP and SCoT markers. J Genet Eng Biotechnol 19:144. https://doi.org/10.1186/s43141-021-00247-7
Yilmaz A, Ciftci V (2021) Genetic relationships and diversity analysis in turkish laurel (Laurus nobilis L.) Germplasm using ISSR and SCoT markers. Mol Bio Rep 48:4537–4547. https://doi.org/10.1007/s11033-021-06474-y
Poczai P, Varga I, Laos M (2013) Advances in plant genetargeted and functional markers: a review. Plant Methods 9:6–37. https://doi.org/10.1186/1746-4811-9-6
Collard BCY, Mackill DJ (2009) Start Codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plant. Plant Mol Biol Rep 27:86–93. https://doi.org/10.1007/s11105-008-0060-5
Singh AK, Rana MK, Singh S, Kumar S, Kumar R, Singh R (2014) CAAT box-derived polymorphism (CBDP): a novel promotertargeted molecular marker for plants. J Plant Biochem Biotechnol 23:175–183. https://doi.org/10.1007/s13562-013-0199-5
Heikrujam M, Kumar J, Agrawal V (2015) Genetic diversity analysis among male and female jojoba genotypes employing gene targeted molecular markers, start codon targeted (SCoT) polymorphism and CAAT box-derived polymorphism (CBDP) markers. Meta Gene 5:90–97. https://doi.org/10.1016/j.mgene.2015.06.001
Heidari P, Etminan A, Azizinezhad R, Khosroshahli M (2017) Genomic variation studies in durum wheat (Triticum turgidum ssp. durum) using CBDP, SCoT and ISSR markers. Indian J Genet Plant Breed 77:379–386. https://doi.org/10.5958/0975-6906.2017.00051.7
Etminan A, Pour-Aboughadareh A, Mohammadi R, Noori A, Ahmadi-Rad A (2018) Applicability of CAAT box-derived polymorphism (CBDP) markers for analysis of genetic diversity in durum wheat. Cereal Res Commun 46:1–9. https://doi.org/10.1556/0806.45.2017.054
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2018) Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst 152:694–703. https://doi.org/10.1080/11263504.2017.1320311
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2017) Assessment of genetic diversity among iranian Triticum germplasm using agro-morphological traits and start codon targeted (SCoT) markers. Cereal Res Commun 45:574–586. https://doi.org/10.1556/0806.45.2017.033
Etminan A, Pour-Aboughadareh A, Mehrabi AA, Shooshtari L, Ahmadi-Rad A, Moradkhani H (2019) Molecular characterization of the wild relatives of wheat using CAAT-box derived polymorphism. Plant Biosyst 153:398–405. https://doi.org/10.1080/11263504.2018.1492993
Agarwal A, Gupta V, Ul Haq S, Jatav PK, Kpthari SL, Kachhwaha S (2019) Assessment of genetic diversity in 29 rose germplasm using SCoT marker. J King Saud Univ Sci 31:780–788. https://doi.org/10.1016/j.jksus.2018.04.022
Fabriki-Ourang S, Karimi H (2019) Assessment of genetic diversity and relationships among Salvia species using gene targeted CAAT box-derived polymorphism markers. J Genet 98:75. https://doi.org/10.1007/s12041-019-1121-2
Ghobadi G, Etminan A, Mehrabi AM, Shooshtari L (2021) Molecular diversity analysis in hexaploid wheat (Triticum aestivum L.) and two Aegilops species (Aegilops crassa and Aegilops cylindrica) using CBDP and SCoT markers. J Genet Eng Biotechnol 19:1–11. https://doi.org/10.1186/s43141-021-00157-8
Yeken MZ, Emiralioğlu O, Çiftçi V, Bayraktar H, Palacioğlu G, Özer G (2022) Analysis of genetic diversity among common bean germplasm by start codon targeted (SCoT) markers. Mol Biol Rep 49:3839–3847. https://doi.org/10.1007/s11033-022-07229-z
Kiani R, Arzani A, Habibi F (2015) Physiology of salinity tolerance in Aegilops cylindrica. Acta Physiol Plant 37:135–145. https://doi.org/10.1007/s11738-015-1881-0
Arabbeigi M, Arzani A, Majidi MM, Kiani R, Tabatabaei BES, Habibi F (2014) Salinity tolerance of Aegilops cylindrical genotypes collected from hyper-saline shores of Uremia Salt Lake using physiological traits and SSR markers. Acta Physiol Plant 36:2243–2251. https://doi.org/10.1007/s11738-014-1602-0
Ahmadi J, Pour-Aboughadareh A, Fabriki Ourang S, Khalili P, Poczai P (2020) Unraveling salinity stress responses in ancestral and neglected wheat species at early growth stage: a baseline for utilization in future wheat improvement programs. Physiol Mol Biol Plants 26:537–549. https://doi.org/10.1007/s12298-020-00768-4
Doyle JJ, Doyle JK (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112. https://doi.org/10.1007/s001220051046
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Earl DA, von Holdt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Atia MAM, El-Moneim DA, Abdelmoneim TK, Reda EH, Shakour ZTA, El-Halawany AM, El-Kashoury ESA, Shams KA, Abdel-Azim NS, Hegazy MEF (2021) Evaluation of genetic variability and relatedness among eight Centaurea species through CAAT-box derived polymorphism (CBDP) and start codon targeted polymorphism (SCoT) markers. Biotechnol Biotechnol Equip 35:1230–1237. https://doi.org/10.1080/13102818.2021.1960891
Shekhawat JK, Rai MK, Shekhawat NS, Kataria V (2018) Exploring genetic variability in Prosopis cineraria using two gene targeted CAAT box-derived polymorphism (CBDP) and start codon targeted (SCoT) polymorphism markers. Mol Biol Rep 45:2359–2367. https://doi.org/10.1007/s11033-018-4400-8
Dumolin-Lapegue S, Demesure B, Fineschi S, Le Corre V, Petit RJ (1997) Phylogeographic structure of white oaks throughout the european continent. Genetics 146:1475–1487. https://doi.org/10.1093/genetics/146.4.1475
Que Y, Pan Y, Lu Y, Yang C, Yang Y, Huang N, Xu L (2014) Genetic analysis of diversity within a chinese local sugarcane germplasm based on start codon targeted polymorphism. Biomed Res Int 2014:1–10. https://doi.org/10.1155/2014/468375
Naghavi MR, Ranjbar M, Zali A, Aghaei MJ, Mardi M, Pirseyedi SM (2009) Genetic diversity of Aegilops crassa and its relationship with Aegilops tauschii and the D genome of wheat. Cereal Res Commun 37:159–167. https://doi.org/10.1556/CRC.37.2009.2.2
Tahernezhad Z, Zamani MJ, Solouki M, Zahravi M, Imamjomeh AA, Jafaraghaei M, Bihamta MR (2010) Genetic diversity of iranian Aegilops tauschii Coss. Using microsatellite molecular markers and morphological traits. Mol Biol Rep 37:3413–3420. https://doi.org/10.1007/s11033-009-9931-6
Wang J, Luo MC, Chen Z, You FM, Wei Y, Zheng Y, Dvorak J (2013) Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol 198:925–937. https://doi.org/10.1111/nph.12164
Qin P, Wang L, Liu K, Mao S, Li Z, Gao S, Shi H, Liu X (2015) Genome wide association study of Aegilops tauschii traits under seedling-stage cadmium stress. The Crop J 3:405–415. https://doi.org/10.1016/j.cj.2015.04.005
Gogniashvili M, Jinjikhadze T, Maisaia I, Akhalkatsi M, Kotorashvili A, Kotaria N, Beridze T, Dudnikov AJ (2016) Complete chloroplast genomes of Aegilops tauschii Coss. And ae. cylindrica host sheds light on plasmon D evolution. Curr Genet 62:791–798. https://doi.org/10.1007/s00294-016-0583-5
Minaei S, Mohammadi SA, Sabouri A, Dadras AR (2022) High genetic diversity in Aegilops tuaschii Coss. Accessions from North Iran as revealed by IRAP and REMAP markers. J Genet Eng Biotechnol 20:86. https://doi.org/10.1186/s43141-022-00363-y
Daneshvar Z, Omidi O, Etminan A, Ebrahimi A (2022) Revealing the genetic diversity and population structure in Aegilops crassa and Aegilops cylindrica species using molecular markers and physio-chemical traits. Cereal Res Commun 50:347–356. https://doi.org/10.1007/s42976-021-00202-9
Randi E, Lucchini V (2002) Detecting rare introgression of domestic dog genes into wild wolf (Canis lupus) populations by bayesian admixture analyses of microsatellite variation. Conserv Genet 3:29–43. https://doi.org/10.1023/A:1014229610646
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Compliance with ethical standards
Authors declared no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bokaei, A.S., Sofalian, O., Sorkhilalehloo, B. et al. Deciphering the level of genetic diversity in some aegilops species using CAAT box-derived polymorphism (CBDP) and start codon target polymorphism (SCoT) markers. Mol Biol Rep 50, 5791–5806 (2023). https://doi.org/10.1007/s11033-023-08488-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08488-0