Abstract
Background
The hepatitis C virus (HCV) is a major cause of illness around the world. HCV genotype 3a is the most prevalent genotype in Thailand. Direct-acting antiviral (DAA) drugs are available for treatment, and these drugs target the NS3, NS5A, and NS5b proteins of HCV. However, HCV variants that are resistant to NS3 protease inhibitors have been found during treatment. This resistance can be naturally occurring or in response to treatment. The purpose of this study is to analyze the codon positions of the main mutation of the partial NS3 gene region of HCV genotype 3a.
Methods
In order to detect mutations and confirm the genotype of HCV genotype 3a, the nucleotide sequencing and amino acid portion of NS3 were analyzed.
Results
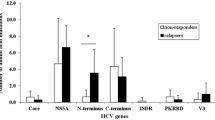
Twenty-six samples were successfully sequenced and clustered within two sub-clades defined as 3a-1 and 3a-2. Through amino acid mutation analysis, the variations were detected at codon positions 122 (3.8%), 132 (84.6%), 168 (100%), 170 (92.3%), 174 (100%), and 175 (100%).
Conclusions
In conclusion, mutations at positions 168, 170, 174, and 175 of the NS3 region are common within the HCV genotype 3a. This information should be useful in the development of effective anti-viral drugs that can successfully treat HCV infection.


Similar content being viewed by others
References
Moradpour D, Penin F, Rice CM (2007) Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463
Nakano T, Lau GMG, Lau GML, Sugiyama M, Mizokami M (2012) An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int 32:339–345
Global progress report on HIV, viral hepatitis and sexually transmitted infections (2021) (https://www.who.int/publications/i/item/9789240027077, accessed 4 Jun 2022.
Irekeola AA, Malek NA, Wada Y, Mustaffa N, Muhamad NI, Shueb RH (2021) Prevalence of HCV genotypes and subtypes in Southeast Asia: A systematic review and meta-analysis. PLoS ONE 16:e0251673
Manns MP, McHutchison JG, Gordon SC et al (2001) Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965
Kowdley KV (2005) Hematologic side effects of interferon and ribavirin therapy. J Clin Gastroenterol 39(1 Suppl):S3–8
European Association for the Study of the Liver, Electronic address, e.e.e. and European Association for the Study of the, L (2018) EASL Recommendations on Treatment of Hepatitis C 2018. J Hepatol 69:461–511
Feld JJ, Jacobson IM, Hézode C et al (2015) Sofosbuvir and Velpatasvir for HCV Genotype 1, 2, 4, 5, and 6 Infection. N Engl J Med 373:2599–2607
Kwo PY, Poordad F, Asatryan A et al (2017) Glecaprevir and pibrentasvir yield high response rates in patients with HCV genotype 1–6 without cirrhosis. J Hepatol 67:263–271
Lawitz E, Poordad F, Gutierrez JA et al (2017) Short-duration treatment with elbasvir/grazoprevir and sofosbuvir for hepatitis C: A randomized trial. Hepatology 65:439–450
Irekeola AA, Ear ENS, Mohd Amin NAZ, Mustaffa N, Shueb RH (2022) Antivirals against HCV infection: the story thus far. J Infect Dev Ctries 16:231–243
Gonzales Zamora JA (2018) Adverse Effects of Direct Acting Antivirals in HIV/HCV Coinfected Patients: A 4-Year Experience in Miami, Florida. Diseases 6:51
Sistayanarain A, Kunthalert D, Vipsoongnern Y (2011) A shift in the hepatitis C virus genotype dominance in blood donor samples from Thailand. Mol Biol Rep 38:4287–4290
Sistayanarain A, Chaiwong S (2017) Molecular characterization of hepatitis C virus genotype 6 subtypes in Thai blood donors. J Microbiol Immunol Infect 50:26–31
Bourliere M, Barberin JM, Rotily M et al (2002) Epidemiological changes in hepatitis C virus genotypes in France evidence in intravenous drug users. J Viral Hepat 9:62–70
Zein NN, Rakela J, Krawitt EL et al (1996) Hepatitis C virus genotypes in the United State: epidemiology, pathogeneicity, and response to interferon therapy. Ann Intern Med 125:634–639
Besse B, Coste-Burel M, Bourgeois N, Feray C, Imbert-Marcille BM, André-Garnier E (2012) Genotyping and resistance profile of hepatitis C (HCV) genotypes 1–6 by sequencing the NS3 protease region using a single optimized sensitive method. J Virol Methods 185:94–100
Pickett BE, Striker R, Lefkowitz EJ (2011) Evidence for separation of HCV subtype 1a into two distinct clades. J Viral Hepat 18:608–618
Vicenti I, Rosi A, Saladini F et al (2012) Naturally occurring hepatitis C virus (HCV) NS3/4A protease inhibitor resistance-related mutations in HCV genotype 1-infected subjects in Italy. J Antimicrob Chemother 67:984–987
Peres-da-Silva A, Almeida AJ, Lampe E (2012) Genetic diversity of NS3 protease from Brazilian HCV isolates and possible implications for therapy with direct-acting antiviral drugs. Mem Inst Oswaldo Cruz 107:254–261
De Luca A, Di Giambenedetto S, Lo Presti A et al (2015) Two Distinct Hepatitis C Virus Genotype 1a Clades Have Different Geographical Distribution and Association With Natural Resistance to NS3 Protease Inhibitors. Open Forum Infect Dis 2:ofv043
Gaudieri S, Rauch A, Pfafferott K et al (2009) Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082
da Silva DL, Nunes HM, Freitas PEB (2021) Natural prevalence of NS3 gene resistance-associated substitutions (RASs) in patients with chronic hepatitis C from the state of Pará/Brazil. Virus Res 292:198251
Wu S, Kanda T, Nakamoto S, Imazeki F, Yokosuka O (2013) Hepatitis C virus protease inhibitor-resistance mutations: our experience and review. World J Gastroenterol 19:8940–8948
Li Z, Chen ZW, Li H, Ren H, Hu P (2017) Prevalence of hepatitis C virus-resistant association substitutions to direct-acting antiviral agents in treatment-naïve hepatitis C genotype 1b-infected patients in western China. Infect Drug Resist 10:377–392
Shepherd SJ, Abdelrahman T, MacLean AR, Thomson EC, Aitken C, Gunson RN (2015) Prevalence of HCV NS3 pre-treatment resistance associated amino acid variants within a Scottish cohort. J Clin Virol 65:50–53
Chen ZW, Li H, Ren H, Hu P (2016) Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep 6:20310
Palanisamy N, Danielsson A, Kokkula C et al (2013) Implications of baseline polymorphisms for potential resistance to NS3 protease inhibitors in Hepatitis C virus genotypes 1a, 2b and 3a. Antiviral Res 99:12–17
Paolucci S, Fiorina L, Piralla A et al (2012) Naturally occurring mutations to HCV protease inhibitors in treatment-naïve patients. Virol J 9:245
Lontok E, Harrington P, Howe A et al (2015) Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology 62:1623–1632
Pawlotsky JM (2016) Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology 151:70–86
Douglas MW, Tay ESE, Wang DS et al (2020) Impact of an Open Access Nationwide Treatment Model on Hepatitis C Virus Antiviral Drug Resistance. Hepatol Commun 4:904–915
Di Stefano M, Faleo G, Farhan Mohamed AM et al (2021) Resistance Associated Mutations in HCV Patients Failing DAA Treatment. New Microbiol 44:12–18
Hoffmann L, Faffe DS, Lima JF et al (2015) No correspondence between resistance mutations in the HCV-NS3 protease at baseline and early telaprevir-based triple therapy. BBA Clin 3:146–151
Sarrazin C (2021) Treatment Failure with DAA Therapy: Importance of Resistance. J Hepatol 74:1472–1482
Acknowledgements
We thank National blood centre, The Thai Red Cross Society, Bangkok, Thailand for providing the samples. The project was supported by National Science, Research and Innovation Fund (NSRF), Year 2021 (R2564B008). Thank you to Mr. Paul Freund of Naresuan Univerisity Writing Clinic (DIALD) for editing assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest.
Ethical approval
Study protocols and procedures were reviewed and approved by the Naresuan University Institutional Review Board (IRB number P10179/63) and the Thai Red Cross Society National Blood Center (Protocal number: 25/2563).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kiattanaphon, A., Vipsoongnern, Y., Kunthalert, D. et al. Partial nonstructural 3 region analysis of hepatitis C virus genotype 3a. Mol Biol Rep 49, 9437–9443 (2022). https://doi.org/10.1007/s11033-022-07803-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07803-5