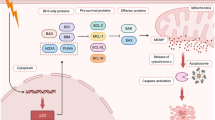
Abstract
The process of cell division plays a vital role in cancer progression. Cell proliferation and error-free chromosomes segregation during mitosis are central events in life cycle. Mistakes during cell division generate changes in chromosome content and alter the balances of chromosomes number. Any defects in expression of TIF1 family proteins, SAC proteins network, mitotic checkpoint proteins involved in chromosome mis-segregation and cancer development. Here we discuss the function of organelles deal with the chromosome segregation machinery, proteins and correction mechanisms involved in the accurate chromosome segregation during mitosis.






Similar content being viewed by others
References
Petry S (2016) Mechanisms of mitotic spindle assembly. Annu Rev Biochem 85:659–683
Westhorpe FG, Straight AF (2014) The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol 7:a015818
Levine MS, Holland AJ (2018) The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev 32:620–638
Potapova T, Gorbsky GJ (2017) The consequences of chromosome segregation errors in mitosis and meiosis. Biology 6
Ricke RM, van Deursen JM (2013) Aneuploidy in health, disease, and aging. J Cell Biol 201:11–21
Wenzel ES, Singh ATK (2018) Cell-cycle checkpoints and aneuploidy on the path to cancer. In Vivo 32:1–5
Barnum KJ, O’Connell MJ (2014) Cell cycle regulation by checkpoints. Methods Mol Biol 1170:29–40
Sansregret L, Swanton C (2017) The role of aneuploidy in cancer evolution. Cold Spring Harb Perspect Med 7:a028373
McVey SL, Cosby JK, Nannas NJ (2021) Aurora B tension sensing mechanisms in the kinetochore ensure accurate chromosome segregation. Int J Mol Sci 22:8818
Yao Y, Dai W (2014) Genomic instability and cancer. J Carcinog Mutagen 5:1000165
Kumar R, Nagpal G, Kumar V, Usmani SS, Agrawal P, Raghava GPS (2019) HumCFS: a database of fragile sites in human chromosomes. BMC Genomics 19:985
Lee JK, Choi YL, Kwon M, Park PJ (2016) Mechanisms and consequences of cancer genome instability: lessons from genome sequencing studies. Annu Rev Pathol 11:283–312
Pikor L, Thu K, Vucic E, Lam W (2013) The detection and implication of genome instability in cancer. Cancer Metastasis Rev 32:341–352
Burrell RA, McGranahan N, Bartek J, Swanton C (2013) The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501:338
Vieira MLC, Santini L, Diniz AL, Munhoz CF (2016) Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 39:312–328
Cortes-Ciriano I, Lee S, Park W-Y, Kim T-M, Park PJ (2017) A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 8:15180–15180
Duijf PHG, Nanayakkara D, Nones K, Srihari S, Kalimutho M, Khanna KK (2019) Mechanisms of genomic instability in breast cancer. Trends Mol Med 25:595–611
Ferguson LR, Chen H, Collins AR, Connell M, Damia G, Dasgupta S, Malhotra M, Meeker AK, Amedei A, Amin A, Ashraf SS, Aquilano K, Azmi AS, Bhakta D, Bilsland A, Boosani CS, Chen S, Ciriolo MR, Fujii H, Guha G, Halicka D, Helferich WG, Keith WN, Mohammed SI, Niccolai E, Yang X, Honoki K, Parslow VR, Prakash S, Rezazadeh S, Shackelford RE, Sidransky D, Tran PT, Yang ES, Maxwell CA (2015) Genomic instability in human cancer: molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Sem Cancer Biol 35:S5–S24
McAvera RM, Crawford LJ (2020) TIF1 proteins in genome stability and cancer. Cancers 12:2094
Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, Béchade G, Van Dorsselaer A, Sanglier-Cianférani S, Hamiche A, Cammas F, Davidson I, Losson R (2011) Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci 108:8212
Pommier RM, Gout J, Vincent DF, Alcaraz LB, Chuvin N, Arfi V, Martel S, Kaniewski B, Devailly G, Fourel G, Bernard P, Moyret-Lalle C, Ansieau S, Puisieux A, Valcourt U, Sentis S, Bartholin L (2015) TIF1γ suppresses tumor progression by regulating mitotic checkpoints and chromosomal stability. Cancer Res 75:4335–4350
Pesenti ME, Weir JR, Musacchio A (2016) Progress in the structural and functional characterization of kinetochores. Curr Opin Struct Biol 37:152–163
Prosser SL, Pelletier L (2017) Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol 18:187–201
Bancroft J, Auckland P, Samora CP, McAinsh AD (2015) Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J Cell Sci 128:171–184
Prosser SL, Pelletier L (2017) Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol 18:187
Vigneron S, Prieto S, Bernis C, Labbé J-C, Castro A, Lorca T (2004) Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15:4584–4596
Diogo V, Teixeira J, Silva PMA, Bousbaa H (2017) Spindle assembly checkpoint as a potential target in colorectal cancer: current status and future perspectives. Clin Colorectal Cancer 16:1–8
Garcia YA, Velasquez EF, Gao LW, Cheung K, Clutario KM, Williams-Hamilton T, Gholkar AA, Whitelegge JP, Torres JZ (2020) Dissection of the spindle assembly checkpoint by proximity proteomics. bioRxiv, 2020.2006.2004.133710
Alhmoud JF, Woolley JF, Moustafa A, Malki MI (2020) DNA damage/repair management in cancers. Cancers 12:1050
Vleugel M, Hoogendoorn E, Snel B, Kops GJ (2012) Evolution and function of the mitotic checkpoint. Dev Cell 23:239–250
Gay S (2018) A novel function for the mitotic checkpoint protein Mad2p in translation. Mol Cell Oncol 5:e1494949–e1494949
Mirkovic M, Hutter LH, Novák B, Oliveira RA (2015) Premature sister chromatid separation is poorly detected by the spindle assembly checkpoint as a result of system-level feedback. Cell Rep 13:469–478
Thompson SL, Bakhoum SF, Compton DA (2010) Mechanisms of chromosomal instability. Curr Biol 20:R285-295
Bakhoum SF, Kabeche L, Murnane JP, Zaki BI, Compton DA (2014) DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov 4:1281
Nicholson JM, Cimini D (2011) How mitotic errors contribute to karyotypic diversity in cancer. In: Gisselsson D (ed) Advances in cancer research. Academic Press, pp 43–75
Gonczy P (2015) Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 15:639–652
Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Eddé B, Bornens M (1998) Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 143:1575–1589
Raff JW, Basto R (2017) Centrosome amplification and cancer: a question of sufficiency. Dev Cell 40:217–218
Conduit PT, Wainman A, Raff JW (2015) Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16:611
Gönczy P, Hatzopoulos GN (2019) Centriole assembly at a glance. J Cell Sci 132:jcs228833
Wang JT, Kong D, Hoerner CR, Loncarek J, Stearns T (2017) Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. eLife 6:e29061
Darling S, Fielding AB, Sabat-Pośpiech D, Prior IA, Coulson JM (2017) Regulation of the cell cycle and centrosome biology by deubiquitylases. Biochem Soc Trans 45:1125–1136
Nam H-J, Naylor R, van Deursen JM (2015) Centrosome dynamics as a source of chromosomal instability. Trends Cell Biol 25:65–73
Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D (2012) Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell 23:401–411
Jusino S, Fernández-Padín FM, Saavedra HI (2018) Centrosome aberrations and chromosome instability contribute to tumorigenesis and intra-tumor heterogeneity. J Cancer Meta Treat 4(8)
Wu Q, Li B, Liu L, Sun S, Sun S (2020) Centrosome dysfunction: a link between senescence and tumor immunity. Signal Transduct Target Therapy 5:107
Cosenza MR, Cazzola A, Rossberg A, Schieber NL, Konotop G, Bausch E, Slynko A, Holland-Letz T, Raab MS, Dubash T, Glimm H, Poppelreuther S, Herold-Mende C, Schwab Y, Krämer A (2017) Asymmetric centriole numbers at spindle poles cause chromosome missegregation in cancer. Cell Rep 20:1906–1920
Lopes CAM, Mesquita M, Cunha AI, Cardoso J, Carapeta S, Laranjeira C, Pinto AE, Pereira-Leal JB, Dias-Pereira A, Bettencourt-Dias M, Chaves P (2018) Centrosome amplification arises before neoplasia and increases upon p53 loss in tumorigenesis. J Cell Biol
LoMastro GM, Holland AJ (2019) The emerging link between centrosome aberrations and metastasis. Dev Cell 49:325–331
Rivera-Rivera Y, Saavedra HI (2016) Centrosome—a promising anti-cancer target. Biologics 10:167–176
Westhorpe FG, Straight AF (2014) The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harbor Perspect Biol 7:015818–015818
Kursel LE, Malik HS (2016) Centromeres. Curr Biol 26:R487–R490
Verdaasdonk JS, Bloom K (2011) Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12:320–332
McKinley KL, Cheeseman IM (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17:16–29
McKinley KL, Cheeseman IM (2015) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17:16
Perpelescu M, Fukagawa T (2011) The ABCs of CENPs. Chromosoma 120:425–446
Westhorpe FG, Straight AF (2013) Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol 25:334–340
Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421
Valdivia M, Hamdouch M, Ortiz K, Astola A (2009) CENPA a genomic marker for centromere activity and human diseases. Curr Genom 10:326–335
Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476:232–235
McKinley KL, Cheeseman IM (2014) Polo-like Kinase 1 Licenses CENP-A Deposition at Centromeres. Cell 158:397–411
Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, Lampson MA, Black BE (2015) CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Sci (New York N Y) 348:699
Hinshaw SM, Makrantoni V, Harrison SC, Marston AL (2017) The kinetochore receptor for the Cohesin loading complex. Cell 171:72-84.e13
Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145:410–422
Huis In ‘t Veld PJ, Jeganathan S, Petrovic A, Singh P, John J, Krenn V, Weissmann F, Bange T, Musacchio A. Molecular basis of outer kinetochore assembly on CENP-T. eLife 5:e21007
Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischböck J, Pentakota S, Keller J, Pesenti ME, Pan D, Vogt D, Wohlgemuth S, Herzog F, Musacchio A (2016) Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 537:249
Foley EA, Kapoor TM (2013) Microtubule attachment and spindle assembly checkpoint signaling at the kinetochore. Nat Rev Mol Cell Biol 14:25–37
Gascoigne KE, Cheeseman IM (2011) Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol 23:102–108
Renda F, Khodjakov A (2021) Role of spatial patterns and kinetochore architecture in spindle morphogenesis. Semin Cell Dev Biol 117:75–85
Musacchio A (2015) The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25:R1002–R1018
Godek KM, Kabeche L, Compton DA (2015) Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol 16:57–64
Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A (2015) CENP-C is a blueprint for constitutive centromere–associated network assembly within human kinetochores. J Cell Biol 210:923–934
Hinshaw SM, Harrison SC (2018) Kinetochore function from the bottom up. Trends Cell Biol 28:22–33
Vicente JJ, Wordeman L (2015) Mitosis, microtubule dynamics and the evolution of kinesins. Exp Cell Res 334:61–69
Chen Y, Hancock WO (2015) Kinesin-5 is a microtubule polymerase. Nat Commun 6:8160
Krenn V, Musacchio A (2015) The aurora B kinase in chromosome Bi-orientation and spindle checkpoint signaling. Front Oncol 5:225
Ji Z, Gao H, Yu H (2015) Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science (New York N Y) 348:1260
Lara-Gonzalez P, Westhorpe FG, Taylor SS (2012) The spindle assembly checkpoint. Curr Biol 22:R966–980
Etemad B, Kops GJ (2016) Attachment issues: kinetochore transformations and spindle checkpoint silencing. Curr Opin Cell Biol 39:101–108
Park I, Lee H-o, Choi E, Lee Y-K, Kwon M-S, Min J, Park P-G, Lee S, Kong Y-Y, Gong G, Lee H (2013) Loss of BubR1 acetylation causes defects in spindle assembly checkpoint signaling and promotes tumor formation. J Cell Biol 202:295
Fujimitsu K, Grimaldi M, Yamano H (2016) Cyclin-dependent kinase 1–dependent activation of APC/C ubiquitin ligase. Science (New York N Y) 352:1121
Kataria M, Yamano H (2019) Interplay between phosphatases and the anaphase-promoting complex/cyclosome in mitosis. Cells 8
Cheng J-M, Liu Y-X (2017) Age-related loss of cohesion: causes and effects. Int J Mol Sci 18
London N, Biggins S (2014) Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 15:736–747
Proudfoot KG, Anderson SJ, Dave S, Bunning AR, Roy S, Bera P, Gupta ML Jr (2019) Checkpoint proteins Bub1 and Bub3 delay anaphase onset in response to low tension independent of microtubule-kinetochore detachment. Cell Rep 27:416-428e414
Bonner MK, Haase J, Swinderman J, Halas H, Miller Jenkins LM, Kelly AE (2019) Enrichment of Aurora B kinase at the inner kinetochore controls outer kinetochore assembly. J Cell Biol 218:3237–3257
Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9:33–46
Bielski CM, Taylor BS (2021) Homing in on genomic instability as a therapeutic target in cancer. Nat Commun 12:3663
Zhu LJ, Pan Y, Chen XY, Hou PF (2020) BUB1 promotes proliferation of liver cancer cells by activating SMAD2 phosphorylation. Oncol Lett 19:3506–3512
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that we have no conflict of interest.
Research involving human and animal participants
The authors declare that the paper did not involve human/animal research.
Informed consent
This paper does not require informed consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jamasbi, E., Hamelian, M., Hossain, M.A. et al. The cell cycle, cancer development and therapy. Mol Biol Rep 49, 10875–10883 (2022). https://doi.org/10.1007/s11033-022-07788-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07788-1