Abstract
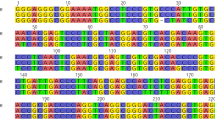
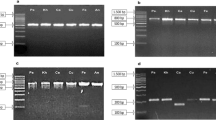
Several species of the Lamiaceae family are the primary source of bioactive aromatic oils and secondary metabolites, having broader applications in the cosmetics, pharmaceuticals, food, confectionery and liquor industries. Due to the scarcity of raw materials and high costs of this family's economically vital species, its products often adulterated to cater to the market's high demand. The present study provides a DNA based approach for identifying different species of this family. Henceforth, the performance of three already proposed barcode loci (matK, trnH-psbA and trnL) was examined for their PCR amplification and species recognition efficacy on various Lamiaceae species and cultivars using three different approaches such as pairwise genetic distance method, BLASTn and phylogenetic tree based on maximum likelihood (ML) analysis. Results illustrate that among all the DNA barcoding loci, matK locus can accurately and efficiently distinguish all the studied species followed by trnH-psbA and trnL. Present investigation may help diminish the illegal trade and events of adulteration of medicinally important plants species in genus Mentha, Ocimum and Plectranthus. This investigation will also help fulfil the scarcity of sequences of barcode loci of these species in the NCBI database. Apart from providing a molecular level reference for identifying processed herbal products, this technique also offers a convenient method for species identification and germplasm conservation of the Lamiaceae family.



Similar content being viewed by others
References
Ahmed SM (2018) Molecular Identification of Lavendula dentata L., Mentha longifolia (L.) HUDS. and Mentha × Piperita by DNA barcodes. Bangladesh J Plant Taxon 25(2):149–157. https://doi.org/10.3329/bjpt.v25i2.39519
Theodoridis S, Stefanaki A, Tezcan M, Aki C, Kokkini S, Vlachonasios KE (2012) DNA barcoding in native plants of the Labiatae (Lamiaceae family from Chios Island (Greece) and the adjacent Cesme-Karaburun Peninsula (Turkey). Mol Ecol Resour 12:620–633. https://doi.org/10.1111/j.1755-0998.2012.03129.x
Khanuja SPS, Shasany AKA, Srivastava A, Kumar S (2000) Assessment of genetic relationships in Mentha species. Euphytica 111:121–125. https://doi.org/10.1023/A:1003829512956
Boren KE, Young DG, Woolley CL, Smith BL, Carlson RE (2015) Detecting essential oil adulteration. J Environ Anal Chem 2:132. https://doi.org/10.4172/2380-2391.1000132
Prakash P, Gupta N (2005) Therapeutic uses of Ocimum sanctum Linn (Tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J Physiol Pharmacol 49:125–131
Christina VLP, Annamalai A (2014) Nucleotide based validation of Ocimum species by evaluating three candidate barcodes of the chloroplast region. Mol Ecol Resour 14:60–68. https://doi.org/10.1111/1755-0998.12167
Tripathi N, Saini N, Tiwari S (2013) Morphological and molecular characterization of endangered medicinal plant species Coleus forskohlii collected from Central India. J Crop Sci Biotech 16:253–261. https://doi.org/10.1007/s12892-013-0083-9
De Mattia F, Bruni I, Galimberti A, Cattaneo F, Casiraghi M, Labra M (2011) A comparative study of different DNA barcoding markers for the identification of some members of Lamiaceae. Food Res Int 44:693–702. https://doi.org/10.1016/j.foodres.2010.12.032
Techen N, Parveen I, Pan Z, Khan IA (2014) DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol 25:103–110. https://doi.org/10.1016/j.copbio.2013.09.010
Thakur VV, Tiwari S, Tripathi N, Tiwari G (2019) Molecular identification of medicinal plants with amplicon length polymorphism using universal DNA barcodes of the atpF–atpH, trnL and trnH–psbA regions. 3Biotech 9:188. https://doi.org/10.1007/s13205-019-1724-6
Thakur VV, Tiwari S, Tripathi N, Tiwari G, Sapre S (2016) DNA barcoding and phylogenetic analyses of Mentha species using rbcL sequences. Ann Phytomed 5(1):59–62
Trindade H (2007) Molecular biology of aromatic plants and spices. Flavour Frag J 25:272–281. https://doi.org/10.1002/ffj.1974
Azizi A, Wagner C, Honermeier B, Wolfgang F (2009) Intraspecific diversity and relationship between subspecies of Origanum vulgare revealed by comparative AFLP and SAMPL marker analysis. Plant Syst Evol 281:151–160. https://doi.org/10.1007/s00606-009-0197-1
Novak J, Lukas B, Bolzer K, Grausgruber-Groger S, Degenhardt J (2008) Identification and characterization of simple sequence repeat markers from a glandular Origanum vulgare expressed sequence tag. Mol Ecol Notes 8:599–601. https://doi.org/10.1111/j.1471-8286.2007.02059.x
Segarra-Moragues JG, Gleiser G (2009) Isolation and characterisation of di and tri nucleotide microsatellite loci in Rosmarinus officinalis (Lamiaceae), using enriched genomic libraries. Conserv Genet 10:571–575. https://doi.org/10.1007/s10592-008-9572-7
Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Philos Trans Ser B 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Tripathi AM, Tyagi A, Kumar A, Singh A, Singh S et al (2013) The internal transcribed spacer (ITS) region and trnhH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE 8(2):e57934. https://doi.org/10.1371/journal.pone.0057934
Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP et al (2005) Land plants and DNA barcodes: short-term and long-term goals. Phil Trans R Soc Lond B Biol Sci 360:1889–1895. https://doi.org/10.1098/rstb.2005.1720
Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD (2007) Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7:135. https://doi.org/10.1186/1471-2148-7-135
CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106:12794–12797. https://doi.org/10.1073/pnas.0905845106
Taberlet P, Gielly L, Pautou G, Bouvet J (2009) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109. https://doi.org/10.1007/BF00037152
Borsch T, Hilu K, Quandt D, Wilde V, Neinhuis C, Barthlott W (2003) Noncoding plastid trnT-trnFsequences reveal a well resolved phylogeny of basal angiosperms. J Evol Biol 16:558–576. https://doi.org/10.1046/j.1420-9101.2003.00577.x
Ridgway K, Duck J, Young J (2003) Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol 3:8. https://doi.org/10.1186/1472-6785-3-8
Tsai L, Yu Y, Hsieh H, Wang J, Linacre A, Lee JC (2006) Species identification using sequences of the trnL intron and the trnL-trnFIGS of chloroplast genome among popular plants in Taiwan. Forensic Sci Int 164:193–200. https://doi.org/10.1016/j.forsciint.2006.01.007
Spaniolas S, Bazakos C, Spano T, Zoghby C, Kalaitzis P (2010) The potential of plastid trnL(UAA) intron polymorphisms for the identification of the botanical origin of plant oils. Food Chem 122:850–856. https://doi.org/10.1016/j.foodchem.2010.02.039
Kress WJ, Erickson DL (2008) DNA barcodes: genes, genomics, and bioinformatics. Proc Natl Acad Sci U S A 105:2761–2762. https://doi.org/10.1073/pnas.0800476105
Ford CS, Ayres KL, Toomey N et al (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159:1–11. https://doi.org/10.1111/j.1095-8339.2008.00938.x
Hollingsworth ML, Clark A, Forrest LL et al (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457. https://doi.org/10.1111/j.1755-0998.2008.02439.x
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Van Velzen R, Weitschek E, Felici G, Bakker FT (2012) DNA barcoding of recently diverged species: relative performance of matching methods. PLoS ONE 7:e30490. https://doi.org/10.1371/journal.pone.0030490
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. https://doi.org/10.1093/molbev/mst024
Guindon S, Dufayard JF, Lefort V, Anisimova M et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Wang Y, Dietrich CH, Zhang Y (2017) Phylogeny and historical biogeography of leafhopper subfamily Evacanthinae (Hemiptera: Cicadellidae) based on morphological and molecular data. Sci Rep 7:45387. https://doi.org/10.1038/srep45387
Erkens RHJ, Cross H, Maas JW, Hoenselaar K, Chatrou LW (2008) Assessment of age and greenness of herbarium specimens as predictors for successful extraction and amplification of DNA. Blumea 53:407–428. https://doi.org/10.3767/000651908X608052
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14. https://doi.org/10.1093/nar/gkl938
Groot DGA, During HJ, Maas JW, Schneider H, Vogel JC (2011) Use of rbcL and trnL-F as a two locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS ONE 6:e16371. https://doi.org/10.1371/journal.pone.001637
Gravendeel B, Eurlings M, Heijerman T (2009) Use of DNA barcoding for host plant identification. Entomol Ber 69:30–35
Tsai CC, Chen CH, Chou CH (2012) DNA bar-codes reveal high levels of morphological plasticity among Rhododendron species (Ericaceae) in Taiwan. Biochem Syst Ecol 40:169–177
Chen CW, Huang YM, Kuo LY, Nguyen QD, Luu HT, Callado JR, Farrar DR, Chiou WL (2012) trnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann Bot. https://doi.org/10.1093/aob/mct004
Pettengill JB, Neel MC (2012) An evaluation of candidate plant DNA barcodes and assignment methods in diagnosing 29 species in the genus Agalinis (Orobanchaceae). Am J Bot 97:1391–1406. https://doi.org/10.3732/ajb.0900176
Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7:e35071. https://doi.org/10.1371/journal.pone.0035071
Newmaster SG, Ragupathy S (2009) Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol Ecol Resour 9:172–180. https://doi.org/10.1111/j.1755-0998.2009.02642.x
Lahaye R, van der Bank M, Bogarin D et al (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105:2923–2928. https://doi.org/10.1073/pnas.0709936105
Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008) Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour 8:480–490. https://doi.org/10.1111/j.1471-8286.2007.02002.x
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374. https://doi.org/10.1073/pnas.0503123102
Gobert V, Moja S, Colson M, Taberlet P (2002) Hybridization in the section Mentha (Lamiaceae) inferred from AFLP markers. Am J Bot 89:2017–2023. https://doi.org/10.3732/ajb.89.12.2017
Tucker AO (1992) The truth about mints. Herb Companion 4:51–52
Cowan RS, Chase MW, Kress WJ, Savolainen V (2006) 300,000 species to identify: problems, progress, and prospects in DNA barcoding of land plants. Taxon 55:611–616. https://doi.org/10.2307/25065638
Paton A, Harley MR, Harley MM (1999) Ocimum: an overview of classification and relationships. In: Hiltunen R, Holm Y (eds) Basil: the genus Ocimum. Harwood, Amsterdam, pp 1–38
Simon JE, Morales MR, Phippen WB, Vieira RF, Hao Z (1999) Basil: a source of aroma compounds and a popular culinary and ornamental herb. In: Janick J (ed) Perspectives on new crops and new uses. ASHS Press, Alexandria, pp 499–505
Waldiaa S, Joshi BC, Pathaka U, Joshi MC (2011) The genus Plectranthus in India and Its chemistry. Chem Biodivers 8:244–252. https://doi.org/10.1002/cbdv.201000048
Funding
ST acknowledges a grant from Madhya Pradesh State Agriculture Marketing Board, Bhopal, India.
Author information
Authors and Affiliations
Contributions
ST and VVT conceived and designed the research. VVT conducted experiments and analyzed the data. NT helped with the experiments and data analysis. VVT and NT wrote the manuscript. ST edited the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thakur, V.V., Tripathi, N. & Tiwari, S. DNA barcoding of some medicinally important plant species of Lamiaceae family in India. Mol Biol Rep 48, 3097–3106 (2021). https://doi.org/10.1007/s11033-021-06356-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06356-3