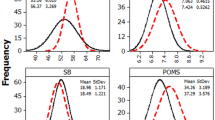
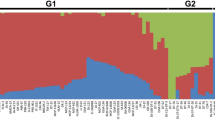
Abstract
Nitrogen (N) is a critical input for plant growth and development. A better understanding of N uptake and utilization is important to develop plant breeding strategies for improving nitrogen use efficiency (NUE). With that objective in mind, we assayed a SNP-genotyped association panel comprising 92 inbred lines for the activities of nitrate reductase (NR), nitrite reductase (NIR), glutamine synthetase (GS) and glutamate synthase (GOGAT). All these enzymes are associated with N assimilation. The experiments were carried out at two levels of N application: no added N (N0) and agrnomically recommened dose (100 kg/ha) of N application (N100). Genome wide association studies (GWAS) helped to identify several marker-trait associations (MTAs), involving chromosomes A01, A06, A08, B02, B04, B05 and B08. These explained high phenotypic variation (up to 32%). Annotation of the genomic region(s) in or around significant SNPs allowed prediction of genes encoding high affinity nitrate transporters, glutamine synthetase 1.3, myb-like transcription factor family protein, bidirectional amino acid transporter 1, auxin signaling F-box 3 and oxidoreductases. This is the first attempt to use GWAS for identification of enzyme QTLs to explain variation for nitrogen assimilation enzymes in Brassica juncea.



Similar content being viewed by others
References
Goel P, Sharma NK, Bhuria M, Sharma V, Chauhan R, Pathania S, Swarnkar MK, Chawla V, Acharya V, Shankar R, Singh AK (2018) Transcriptome and co-expression network analyses identify key genes regulating nitrogen use efficiency in Brassica juncea L. Sci Rep 8:7451
Miller AJ, Cookson SJ, Smith SJ, Wells DM (2001) The use of microelectrodes to investigate compartmentation and the transport of metabolized inorganic ions in plants. J Exp Bot 52:541–549
Sylvester-Bradley R, Kindered DR (2009) Analysing nitrogen responses of cereals to prioritize routes to the improvement of nitrogen use efficiency. J Exp Bot 60:1939–1951
Makino A (2011) Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol 155:125–129
Sinclair TR, Rufty TW (2012) Nitrogen and water resources commonly limit crop yield increases, not necessarily plant genetics. Glob Food Sec 1:94–98
Sebilo M, Mayer B, Nicolardot B, Pinay G, Mariotti A (2013) Long-term fate of nitrate fertilizer in agricultural soils. Proc Nat Acad Sci 110:18185–18189
Dechorgnat J, Francis KL, Dhugga KS, Rafalski JA, Tyerman SD, Kaiser BN (2018) Root ideotype influences nitrogen transport and assimilation in maize. Front Plant Sci 9:531. https://doi.org/10.3389/fpls.2018.00531
Leleu O, Vuylstecker C, Tetu JF, Degrande D, Champolivier L, Rambour S (2000) Effect of two contrasted N fertilizations on rapeseed growth and nitrate metabolism. Plant Physiol Biochem 38:639–645
Chamorro AM, Tamagno LN, Bezus R, Sarandon SJ (2002) Nitrogen accumulation, partition and nitrogen use efficiency in canola under different nitrogen availabilities. Commun Soil Sci Plan 33:493–504
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105:1141–1157
Glass AD, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, Vidmar JJ (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53:855–864
Gupta S, Atwal AK, Kumar H, Sardana V, Sangha MK, Kumar N, Banga SS (2013) Characterizing oilseed Brassica germplasm for traits associated with nitrogen use efficiency using biplot analysis. Crop Improv 40:34–43
Fischer K, Barbier GG, Hecht HJ, Mendel RR, Campbell WH, Schwarz G (2005) Structural basis of eukaryotic nitrate reduction: crystal structures of the nitrate reductase active site. Plant Cell 17:1167–1179
Kouadiao JY, Kouakou HT, Kone M, Zouzou M, Anno PA (2007) Optimum conditions for cotton nitrate reductase extraction and activity measurement. Afr J Biotechnol 6:923–928
Cao Y, Fan XR, Sun S, Xu G, Hu J, Shen QR (2008) Effect of nitrate on activities and transcript levels of nitrate reductase and glutamine synthetase in rice. Pedosphere 18:664–673
Rosales EP, Iannone MF, Groppa MD, Benavides MP (2011) Nitric oxide inhibits nitrate reductase activity in wheat leaves. Plant Physiol Biochem 49:124–130
Miflin BJ (1974) The location of nitrite reductase and other enzymes related to amino acid biosynthesis in the plastids of root and leaves. Plant Physiol 54:550–555
Wray JL (1993) Molecular biology, genetics and regulation of nitrite reduction in higher plants. Physiol Plant 89:607–612
Cren M, Hirel B (1999) Glutamine synthetase in higher plants: regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol 40:1187–1193
Miflin BJ, Habash DJ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53:979–987
Cai H, Zhou Y, Xiao J, Li X, Zhang Q, Lian X (2009) Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep 28:527–537
Suzuki A, Knaff DB (2005) Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth Res 83:91–217
Esposito S, Guerriero G, Vona V, Rigano VDM, Carfagna S, Rigano C (2005) Glutamate synthase activities and protein changes in relation to nitrogen nutrition in barley: the dependence on different plastidic glucose-6P dehydrogenase isoforms. J Exp Bot 56:55–64
Tischner R (2000) Nitrate uptake and reduction in higher and lower plants. Plant Cell Environ 23:1005–1024
Mokhele B, Zhan X, Yang G, Zhang X (2012) Review: nitrogen assimilation in crop plants and its affecting factors. Can J Plant Sci 92:399–405
Zhang N, Gibon Y, Wallace JG, Lepak N, Li P, Dedow L, Chen C, So YS, Kremling K, Bradbury PJ, Brutnell T, Stitt M, Buckler ES (2015) Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiol 168:575–583
Mackay L, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends Plant Sci 12:57–63
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Waugh R, Jannink JL, Muehlbauer GJ, Ramsay L (2009) The emergence of whole genome association scans in barley. Curr Opin Plant Biol 12:218–222
Ingvarsson PK, Nathaniel RS (2011) Association genetics of complex traits in plants. New Phytol 189:909–922
Wei X (2011) Genome-wide association analysis and phenotypic study of nitrogen use efficiency in Arabidopsis thaliana. Master Thesis, Plant Genetics, Wageningen University, Wageningen, The Netherlands
Chen GF, Wu RG, Li DM, Yu HX, Deng Z, Tian JC (2017) Genome wide association study for seeding emergence and tiller number using SNP markers in an elite winter wheat population. J Genet 96:177–186
Matthies IE, Weise S, Forster J, Korzun V, Stein N (2013) Nitrogen-metabolism related genes in barley - haplotype diversity, linkage mapping and associations with malting and kernel quality parameters. BMC Genet 14:77
Sinha SK, Sevanthi VAM, Chaudhary S, Tyagi P, Venkadesan SK, Rani M, Mandal PK (2018) Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic nitrogen starvation reveals differences in chloroplast and starch metabolism-related genes. Genes 9:206
Morrison KM, Simmons SJ, Stapleton AE (2010) Loci controlling nitrate reductase activity in maize: ultraviolet-B signaling in aerial tissues increases nitrate reductase activity in leaf and root when responsive alleles are present. Physiol Plant 140:334–341
Goel P, Singh AK (2015) Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS ONE 10:e0143645
Jaworski EG (1971) Nitrate reductase in intact plant tissue. Biochem Biophys Res Commun 43:1274–1279
Verner JE, Ferari TE (1971) Intact tissue assay for nitrite reductase in barley aleurone layers. Plant Physiol 47:790–794
Mohanty B, Fletcher JS (1980) Ammonium influence on nitrogen assimilating enzymes and protein accumulation in suspension cultures of Paul’s Scarlet rose. Physiol Plant 48:453–459
Bulen WA (1956) The isolation and characterization of glutamate dehydrogenase from corn leaves. Arch Biochem Biophys 62:178–183
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Duitama J, Quintero JC, Cruz DF, Quintero C, Hubmann G, Foulquie-Moreno MR, Verstrepen KJ, Thevelein JM, Tohme J (2014) An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res 42(6):e44. https://doi.org/10.1093/nar/gkt1381
Roshyara NR, Scholz M (2014) fcgene: a versatile tool for processing and transforming SNP datasets. PLoS ONE 9:e97589
Browning BL, Browning SR (2007) Efficient multilocus association mapping for whole genome association studies using localized haplotype clustering. Genet Epidemiol 31:365–375
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) Software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Wang Q, Tian F, Pan Y, Buckler ES, Zhang Z (2014) A SUPER powerful method for genome wide association study. PLoS ONE 9:e107684
Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acid Res 36:3420–3435
Malagoli P, Laine P, Rossato L, Ourry A (2005) Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. I. Global N flows between vegetative and reproductive tissues in relation to leaf fall and their residual N. Ann Bot 95:853–861
Malagoli P, Laine P, Rossato L, Ourry A (2005) Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. II. An 15N-labelling-based simulation model of N partitioning between vegetative and reproductive tissues. Ann Bot 95:1187–1198
Tilsner J, Kassner N, Struck C, Lohaus G (2005) Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 221:328–338
Gombert J, Etienne P, Ourry F, Le Dily F (2006) The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J Exp Bot 57:1949–1956
Avice JC, Etienne P (2014) Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J Exp Bot 65:3813–3824
Bussi C, Gojon A, Passama L (1997) In situ nitrate reductase activity in leaves of adult peach trees. J Hortic Sci 72:347–353
Sanchez E, Rivero RM, Ruiz JM, Romero L (2004) Yield and biosynthesis of nitrogenous compounds in fruits of green bean (Phaseolus vulgaris L cv. Strike) in response to increasing N fertilization. J Sci Food Agric 84:575–580
Ahmad A, Khan I, Anjum NA, Diva I, Abdin MZ, Iqbal M (2005) Effect of timing of sulphur fertilizer application on growth and yield of rapeseed (Brassica campestris L.). J Plant Nut 28:1049–1059
Shangguan ZP (2007) Effects of nitrogen application rate on nitrate reductase activity, nitric oxide content and gas exchange in winter wheat leaves. Chin J Appl Ecol 18:1447–1452
Reis AR, Favarin JL, Gallo LA, Malavolta E, Moraes MF, Junior JL (2009) Nitrate reductase and glutamine synthetase activity in coffee leaves during fruit development. Rev Bras Cienc Solo 33:315–324
Chandna R, Hakeem KR, Khan F, Ahmad A, Iqbal M (2012) Variability of nitrogen uptake and assimilation among N-efficient and N-inefficient wheat (Triticum aestivum L.) genotypes. J Plant Interact 7:367–375
Fan X, Gordon-Weeks R, Shen Q, Miller AJ (2006) Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J Exp Bot 57:1333–1340
Obara M, Kajiura M, Fukuta Y, Yano M, Hayashi M, Yamaya T, Sato T (2001) Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J Exp Bot 52:1209–1217
Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Nat Acad Sci 107:4477–4482
Kim Y, Kim H, Son H, Choi GJ, Kim JC, Lee YW (2014) MYT3, a myb-like transcription factor, affects fungal development and pathogenicity of Fusarium graminearum. PLoS ONE 9:e94359. https://doi.org/10.1371/journal.pone.0094359
Wipf D, Loque D, Lalonde S, Wolf BF (2012) Amino acid transporter inventory of the Selaginella genome. Front Plant Sci 3:36
Besnard J, Pratelli R, Zhao C, Sonawala U, Collakova E, Pilot G, Okumoto S (2016) UMAMIT14 is an amino acid exporter involved in phloem unloading in Arabidopsis roots. J Exp Bot 67:6385–6397
Bernard SM, Habash DZ (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 182:608–620
Krapp A, Fraisier V, Scheible WR, Quesada A, Gojon A, Stitt M, Caboche M, Daniel-Vedele F (1998) Expression studies of Nrt 2:1Np, a putative high-affinity nitrate transporter: evidence for its role in nitrate uptake. Plant J 14:723–731
Vidmar JJ, Zhuo D, Siddiqi MY, Schoerring JK, Touraine B, Glass AD (2000) Regulation of high affinity nitrate transporter genes and high affinity nitrate influx by nitrogen pools in plant roots. Plant Physiol 123:307–318
Nazoa P, Vidmar JJ, Tranbarger T, Mouline K, Damiani I, Tillard P, Zhuo D, Glass ADM, Touraine B (2003) Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol Biol 52:689–703
Acknowledgements
The studies were financially supported by the Department of Biotechnology, Government of India in the form of Centre of Excellence and Innovation in Biotechnology “Germplasm enhancement for crop architecture and defensive traits in Brassica juncea L. Czern. and Coss”. SSB also acknowledges salary support from Indian Council of Agricultural Research under ICAR National Professor Chair.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, S., Akhatar, J., Kaur, P. et al. Genetic analyses of nitrogen assimilation enzymes in Brassica juncea (L.) Czern & Coss. Mol Biol Rep 46, 4235–4244 (2019). https://doi.org/10.1007/s11033-019-04878-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04878-5