Abstract
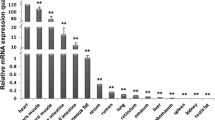
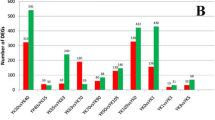
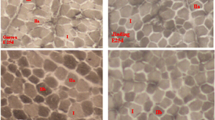
The FABP (adipocyte fatty acid-binding protein) genes play an important role in intracellular fatty acid transport and considered to be candidate genes for fatness traits in domestic animal. In this study, we cloned the cDNA sequences of goat FABP family genes and their expression patterns were detected by semi-quantitative RT-PCR and quantitative real time RT-PCR. Expression analysis showed that goat FABP1 gene was predominantly expressed in liver, kidney and large intestine. While FABP4 was widely expressed in many tissues with a high expression level was observed in the fat, skeletal muscle, stomach and lung. Notably, FABP2 gene was expressed specifically in small intestine. Moreover, goat FABP3 was expressed at 60 day with the highest level, then significantly (p < 0.01) decreased at the 90 day. No significant expression differences were observed in longissimus dorsi muscles among 3 day, 30 day and 60 day. Goat FABP4 was expressed at 3 day with the lowest level, then significantly (p < 0.01) increased to a peak at the 60 day. In addition, a significant relationship between FABP3 mRNA expression levels and intramuscular fat (IMF) content was observed. These results suggest that the FABP3 and FABP4 may be important genes for meat quality and provides useful information for further studies on their roles in skeletal muscle IMF deposit.



Similar content being viewed by others
References
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C et al (2010) Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal 4:303–319
Ono T (2005) Studies of the FABP family: a retrospective. Mol Cell Biochem 277:1–6
Gerbens F, de Koning DJ, Harders FL, Meuwissen TH, Janss LL et al (2000) The effect of adipocyte and heart fatty acid-binding protein genes on intramuscular fat and backfat content in Meishan crossbred pigs. J Anim Sci 78:552–559
Gerbens F, Verburg FJ, Van Moerkerk HT, Engel B, Buist W et al (2001) Associations of heart and adipocyte fatty acid-binding protein gene expression with intramuscular fat content in pigs. J Anim Sci 79:347–354
Perez-Enciso M, Clop A, Noguera JL, Ovilo C, Coll A et al (2000) A QTL on pig chromosome 4 affects fatty acid metabolism: evidence from an Iberian by Landrace intercross. J Anim Sci 78:2525–2531
Cho KH, Kim MJ, Jeon GJ, Chung HY (2011) Association of genetic variants for FABP3 gene with back fat thickness and intramuscular fat content in pig. Mol Biol Rep 38:2161–2166
Han X, Jiang T, Yang H, Zhang Q, Wang W et al (2012) Investigation of four porcine candidate genes (H-FABP, MYOD1, UCP3 and MASTR) for meat quality traits in Large White pigs. Mol Biol Rep 39:6599–6605
Barendse W, Bunch RJ, Thomas MB, Harrison BE (2009) A splice site single nucleotide polymorphism of the fatty acid binding protein 4 gene appears to be associated with intramuscular fat deposition in longissimus muscle in Australian cattle. Anim Genet 40:770–773
Jurie C, Cassar-Malek I, Bonnet M, Leroux C, Bauchart D et al (2007) Adipocyte fatty acid-binding protein and mitochondrial enzyme activities in muscles as relevant indicators of marbling in cattle. J Anim Sci 85:2660–2669
Lee SH, van der Werf JH, Park EW, Oh SJ, Gibson JP et al (2010) Genetic polymorphisms of the bovine fatty acid binding protein 4 gene are significantly associated with marbling and carcass weight in Hanwoo (Korean Cattle). Anim Genet 41:442–444
Shin SC, Heo JP, Chung ER (2012) Genetic variants of the FABP4 gene are associated with marbling scores and meat quality grades in Hanwoo (Korean cattle). Mol Biol Rep 39:5323–5330
Wang Q, Guan T, Li H, Bernlohr DA (2009) A novel polymorphism in the chicken adipocyte fatty acid-binding protein gene (FABP4) that alters ligand-binding and correlates with fatness. Comp Biochem Physiol B: Biochem Mol Biol 154:298–302
Ye MH, Chen JL, Zhao GP, Zheng MQ, Wen J (2010) Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-You chicken. Anim Biotechnol 21:14–24
You X, Liu Y, Jiang X, Du H, Liu Z et al (2009) Relationships between single nucleotide polymorphisms of the H-FABP gene and slaughter and meat quality traits in chicken. Biochem Genet 47:511–520
Shi H, Wang Q, Wang Y, Leng L, Zhang Q et al (2010) Adipocyte fatty acid-binding protein: an important gene related to lipid metabolism in chicken adipocytes. Comp Biochem Physiol B: Biochem Mol Biol 157:357–363
Shi H, Wang Q, Zhang Q, Leng L, Li H (2010) Tissue expression characterization of chicken adipocyte fatty acid-binding protein and its expression difference between fat and lean birds in abdominal fat tissue. Poult Sci 89:197–202
Li L, Jiang J, Wang L, Zhong T, Chen B et al (2013) Expression patterns of peroxisome proliferator-activated receptor gamma 1 versus gamma 2, and their association with intramuscular fat in goat tissues. Gene 528:195–200
Hovenier R, Brascamp EW, Kanis E, van der Werf JH, Wassenberg AP (1993) Economic values of optimum traits: the example of meat quality in pigs. J Anim Sci 71:1429–1433
Chmurzynska A (2006) The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet 47:39–48
Storch J, Thumser AE (2010) Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem 285:32679–32683
Storch J, McDermott L (2009) Structural and functional analysis of fatty acid-binding proteins. J Lipid Res 50(Suppl):S126–S131
Newberry EP, Kennedy SM, Xie Y, Luo J, Crooke RM et al (2012) Decreased body weight and hepatic steatosis with altered fatty acid ethanolamide metabolism in aged L-Fabp -/- mice. J Lipid Res 53(4):744–754
Baier LJ, Sacchettini JC, Knowler WC, Eads J, Paolisso G et al (1995) An amino acid substitution in the human intestinal fatty acid binding protein is associated with increased fatty acid binding, increased fat oxidation, and insulin resistance. J Clin Invest 95:1281–1287
Lagakos WS, Gajda AM, Agellon L, Binas B, Choi V et al (2011) Different functions of intestinal and liver-type fatty acid-binding proteins in intestine and in whole body energy homeostasis. Am J Physiol Gastrointest Liver Physiol 300:G803–G814
Tyra M, Ropka-Molik K, Eckert R, Piorkowska K, Oczkowicz M (2011) H-FABP and LEPR gene expression profile in skeletal muscles and liver during ontogenesis in various breeds of pigs. Domest Anim Endocrinol 40:147–154
Michal JJ, Zhang ZW, Gaskins CT, Jiang Z (2006) The bovine fatty acid binding protein 4 gene is significantly associated with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Anim Genet 37:400–402
Toplu HD, Goksoy EO, Nazligul A, Kahraman T (2013) Meat quality characteristics of Turkish indigenous Hair goat kids reared under traditional extensive production system: effects of slaughter age and gender. Trop Anim Health Prod 45:1297–1304
Di Luccia A, Satriani A, Barone CM, Colatruglio P, Gigli S et al (2003) Effect of dietary energy content on the intramuscular fat depots and triglyceride composition of river buffalo meat. Meat Sci 65:1379–1389
Wu T, Zhang Z, Yuan Z, Lo LJ, Chen J et al (2013) Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and landrace pigs. PLoS One 8:e53181
Samulin J, Berget I, Lien S, Sundvold H (2008) Differential gene expression of fatty acid binding proteins during porcine adipogenesis. Comp Biochem Physiol B: Biochem Mol Biol 151:147–152
Gardan D, Louveau I, Gondret F (2007) Adipocyte- and heart-type fatty acid binding proteins are both expressed in subcutaneous and intramuscular porcine (Sus scrofa) adipocytes. Comp Biochem Physiol B: Biochem Mol Biol 148:14–19
Vergara A, Franzese M, Merlino A, Vitagliano L, Verde C et al (2007) Structural characterization of ferric hemoglobins from three antarctic fish species of the suborder notothenioidei. Biophys J 93:2822–2829
Damon M, Louveau I, Lefaucheur L, Lebret B, Vincent A et al (2006) Number of intramuscular adipocytes and fatty acid binding protein-4 content are significant indicators of intramuscular fat level in crossbred Large White x Duroc pigs. J Anim Sci 84:1083–1092
Reiter SS, Halsey CH, Stronach BM, Bartosh JL, Owsley WF et al (2007) Lipid metabolism related gene-expression profiling in liver, skeletal muscle and adipose tissue in crossbred Duroc and Pietrain Pigs. Comp Biochem Physiol Part D Genomics Proteomics 2:200–206
Acknowledgments
This work was supported by the Grants from Scientific Researcher Fund of Sichuan Provincial Education Department (10ZA049), and the national Spark key Program (2012GA810001), and Sichuan Province Science and Technology Support Program (2011NZ0003, 2011NZ0099-36).
Author information
Authors and Affiliations
Corresponding author
Additional information
Linjie Wang and Li Li have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2014_3759_MOESM1_ESM.rtf
Fig. S1 Multiple amino acid sequence alignments of goat, human, mouse and pig FABP1, FABP2, FABP3 and FABP4 genes. Protein sequences were obtained from GenBank: NP_001434.1 (FABP1-Homo sapiens); NP_001004046.1 (FABP1-Sus scrofa); NP_059095.1 (FABP1-Mus musculus); NP_032006.1 (FABP2-Mus musculus); NP_000125.2 (FABP2-Homo sapiens); NP_001026950.1 (FABP2-Sus scrofa); NP_004093.1 (FABP3-Homo sapiens); NP_034304.1 (FABP3-Mus musculus); NP_001093401.1 (FABP3-Sus scrofa); NP_001433.1 (FABP4-Homo sapiens); NP_077717.1 (FABP4-Mus musculus); NP_001002817.1 (FABP4-Sus scrofa). Conserved amino acids are highlighted. The putative lipocalin and fatty acid binding protein signatures in their central portion underlined. (RTF 157 kb)
Rights and permissions
About this article
Cite this article
Wang, L., Li, L., Jiang, J. et al. Molecular characterization and different expression patterns of the FABP gene family during goat skeletal muscle development. Mol Biol Rep 42, 201–207 (2015). https://doi.org/10.1007/s11033-014-3759-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3759-4