Abstract
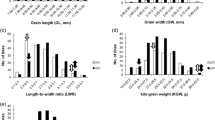
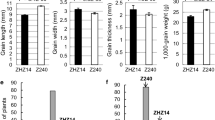
Grain shape is a key factor that influences both grain yield and quality of rice (Oryza sativa L.). In order to elucidate the genetic basis of grain shape, an F2 and derived F2:3 populations were developed from a cross between Guangzhan63-4S (GZ63-4S) and TGMS29, and used in a quantitative trait locus (QTL) analysis. A total of thirty-six QTLs were identified for grain shape and weight, and the phenotypic variance explained by each QTL ranged from 0.88 to 35.70%. Given that most of the QTLs were detected in only one population, a BC1F2 and derived BC1F2:3 populations were constructed with GZ63-4S as the recurrent parent to further verify the QTLs. Seventeen QTLs were validated in at least one population. qGL1.3, the major QTL for grain length in both the F2 and F2:3 populations, contributed 41.16% of the variation in grain length and 35.96% of the variation in 1000-grain weight in a random BC1F3 population, and thus was selected for further fine mapping. Based on progeny tests of 31 recombinants, qGL1.3 was delimited to a region of about 350 kb on chromosome 1. These results could lay the foundation for map-based cloning of qGL1.3 and provide genes or QTLs for breeding rice lines with high grain yield and quality.






Similar content being viewed by others
References
Bai XF, Luo LJ, Yan WH, Kovi MR, Zhan W, Xing YZ (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet 11:16
Che RH, Tong HN, Shi BH et al (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2:15195
Chen HD, Xie WB, He H, Yu H, Chen W, Li J, Yu R, Yao Y, Zhang W, He Y, Tang X, Zhou F, Deng XW, Zhang Q (2014) A high-density SNP genotyping array for rice biology and molecular breeding. Mol Plant 7:541–553
Duan PG, Ni S, Wang JM, Zhang BL, Xu R, Wang YX, Chen HQ, Zhu XD, Li YH (2015) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants 2:15203
Duan PG, Xu JS, Zeng DL, Zhang B, Geng M, Zhang G, Huang K, Huang L, Xu R, Ge S, Qian Q, Li Y (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10:685–694
Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14:133–139
Hu J, Wang YX, Fang YX, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, Zhu L, Dong G, Guo L, Zeng D, Zhang G, Xie L, Xiong G, Li J, Qian Q (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465
Hu ZJ, Lu SJ, Wang MJ, He H, Sun L, Wang H, Liu XH, Jiang L, Sun JL, Xin X, Kong W, Chu C, Xue HW, Yang J, Luo X, Liu JX (2018) A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant 11:736–749
Huang RY, Jiang LR, Zheng JS, Wang TS, Wang HC, Huang YM, Hong ZL (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci 18:218–226
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45:707–711
Liu JF, Chen J, Zheng XM et al (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3:17043
Liu Q, Han RX, Wu K et al (2018) G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat Commun 9:852
Margarido GRA, Souza AP, Garcia AAF (2010) OneMap: software for genetic mapping in outcrossing species. Hereditas 144:78–79
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22:1666–1680
Qi L, Sun Y, Li J, Su L, Zheng XM, Wang XN, Li KM, Yang QW, Qiao WH (2017) Identification of QTLs for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breed Sci 67:472–482
Rasheed A, Hao YF, Xia XC, Khan A, Xu Y, Varshney RK, He Z (2017) Crop breeding chips and genotyping platforms: progress, chanllenges, and perspectives. Mol Plant 10:1047–1064
Singh N, Jayaswal PK, Panda K et al (2015) Single-copy gene based 50K SNP chip for genetic studies and molecular breeding in rice. Sci Rep 5:11600
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Sun PY, Zhang WH, Wang YH, He Q, Shu F, Liu H, Wang J, Wang JM, Yuan LP, Deng HF (2016) OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol 58:836–847
Wang SC, Christopher JB, Zeng ZB (2012a) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wang SK, Wu K, Yuan QB et al (2012b) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44(8):950–954
Wang SK, Li S, Liu Q et al (2015a) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Wang YX, Xiong GS, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, Ye W, Meng X, Liu R, Chen H, Jing Y, Wang Y, Zhu X, Li J, Qian Q (2015b) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Xia D, Zhou H, Liu RJ, Dan W, Li P, Wu B, Chen J, Wang L, Gao G, Zhang Q, He Y (2018) GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol Plant 11:754–756
Xing YZ, Zhang QF (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Yang WN, Guo ZL, Huang CL et al (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5:5087
Yu HH, Xie WB, Li J, Zhou FS, Zhang QF (2014) A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol J 12:28–37
Zhao KY, Tung C, Eizenga GC et al (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2:467
Zhao D, Li PB, Wang LQ, Sun L, Xia D, Luo LJ, Gao GJ, Zhang QL, He YQ (2017) Genetic dissection of large grain shape in rice cultivar ‘Nanyangzhan’ and validation of a grain thickness QTL (qGT3.1) and a grain length QTL (qGL3.4). Mol Breed 37:42
Zhou H, Zhou M, Yang YZ et al (2014) RNase ZS1 processes Ub L40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun 5:4884
Funding
This work was supported by grants from the National Program on R&D of Transgenic Plants (2016ZX08009003-004) and the National Natural Science Foundation (91935303), and earmarked fund for Agriculture Research System (CARS-01-03) in China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Data 1
Primers used in this study. (XLSX 33 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Hou, J., Li, P. et al. Genetic dissection and validation of QTLs for grain shape and weight in rice and fine mapping of qGL1.3, a major QTL for grain length and weight. Mol Breeding 39, 170 (2019). https://doi.org/10.1007/s11032-019-1079-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1079-z