Abstract
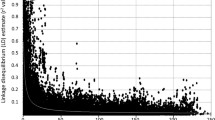
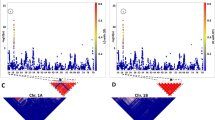
In plant species, construction of framework linkage maps to facilitate quantitative trait loci mapping and molecular breeding has been confined to experimental mapping populations. However, development and evaluation of these populations is detached from breeding efforts for cultivar development. In this study, we demonstrate that dense and reliable linkage maps can be constructed using extant breeding populations derived from a large number of crosses, thus eliminating the need for extraneous population development. Using 565 segregating F1 progeny from 28 four-way cross breeding populations, a linkage map of the hexaploid wheat genome consisting of 3,785 single nucleotide polymorphism (SNP) loci and 22 simple sequence repeat loci was developed. Map estimation was facilitated by application of mapping algorithms for general pedigrees implemented in the software package CRI-MAP. The developed linkage maps showed high rank-order concordance with a SNP consensus map developed from seven mapping studies. Therefore, the linkage mapping methodology presented here represents a resource efficient approach for plant breeding programs that enables development of dense linkage maps “on the fly” to support molecular breeding efforts.











Similar content being viewed by others
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Hettner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113(8):1409–1420
Akhunov E, Chao S, Catana V, See D, Brown-Guedira G, Akhunova A, Dubcovsky J, Cavanagh C, Hayden M (2011) New tools for wheat genetics and breeding: genome-wide analysis of SNP variation. In: McIntosh R (ed) Proceedings of the BGRI 2011 tech. work. Borlaug Global Rust Initiative, St. Paul, MN, pp 92–97
Allen AM, Barker GLA, Berry ST, Coghill JA, Gwilliam R, Kirby S, Robinson P, Brenchley RC, D’Amore R, McKenzie N, Waite D, Hall A, Bevan M, Hall N, Edwards KJ (2011) Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J 9(9):1086–1099
Bandillo N, Raghavan C, Muyco P, Sevilla MAL, Lobina IT, Dilla-Ermita CJ, Tung C-W, McCouch S, Thomson M, Mauleon R, Singh RK, Gregorio G, Rodoña E, Leung H (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6:11
Beavis WD (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex trait. CRC Press, Boca Raton, pp 145–162
Bowers JE, Bachlava E, Brunick RL, Rieseberg LH, Knapp SJ, Burke JM (2012) Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 2(7):721–729
Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol 11(2):215–221
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kianian S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, Lopez da Silva M, Bockelman H, Talber L, Anderson J, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell M, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110(20):8057–8062
Cheema J, Dicks J (2009) Computational approaches and software tools for genetic linkage map estimation in plants. Brief Bioinform 10(6):595–608
Collard B, Mace E, McPhail M et al (2009) How accurate are the marker orders in crop linkage maps generated from large marker datasets? Crop Pasure Sci 52:362–372
Crepieux S, Lebreton C, Servin B, Charmet G (2004) Quantitative trait loci (QTL) detection in multicross inbred designs: recovering QTL identical-by-descent status information from marker data. Genetics 168(3):1737–1749
Crepieux S, Lebreton C, Flament P, Charmet G (2005) Application of a new IBD-based QTL mapping method to common wheat breeding population: analysis of kernel hardness and dough strength. Theor Appl Genet 111(7):1409–1419
Deschamps S, Llaca V, May GD (2012) Genotyping-by-sequencing in plants. Biology (Basel) 1(3):460–483
Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3(1):43–52
Goldstein DR, Zhao H, Speed TP (1997) The effects of genotyping errors and interference on estimation of genetic distance. Hum Hered 47:86–100
Green P, Falls K, Crooks S (1990) Documentation for CRI-MAP, version 2.4. www.animalgenome.org. (Accessed 7 Jan 2014)
Holland JB (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10(2):156–161
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10(7):826–839
Jansen RC, Jannink JL, Beavis WD (2003) Mapping quantitative trait loci in plant breeding populations: use of parental haplotype sharing. Crop Sci 43(3):829–834
Karakousis A, Langridge P (2003) A high-throughput plant DNA extraction method for marker analysis. Plant Mol Biol Rep 21(1):95
Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J, Caig V, Evers M, Heller-Uszynska K, Uszynski G, Cayla C, Patarapuwadol S, Xia L, Yang S, Thomson B (2003) The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In: Proceedings of the international congress in wake double helix from green revolution to gene revolution, Avenue Media, Bologna, Italy, pp 443–461
Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84(8):2363–2367
Morrell PL, Buckler ES, Ross-Ibarra J (2012) Crop genomics: advances and applications. Nat Rev Genet 13(2):85–96
Qin H, Guo W, Zhang Y-M, Zhang T (2008) QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet 117(6):883–894
Rakshit S, Rakshit A, Patil JV (2012) Multiparent intercross populations in analysis of quantitative traits. J Genet 91(1):111–117
Rosyara UR, Gonzalez-Hernandez JL, Glover KD, Gedye KR, Stein JM (2009) Family-based mapping of quantitative trait loci in plant breeding populations with resistance to Fusarium head blight in wheat as an illustration. Theor Appl Genet 118(8):1617–1631
Trebbi D, Maccaferri M, Giuliani S, Sorensen A, Sanguineti MC, Massi A, Tuberosa R (2008) Development of a multi-parental (four-way cross) mapping population for multi-allelic QTL analysis in durum wheat. In: Appels R, Eastwood R, Lagudah E et al (eds) 11th proceedings of the international wheat genetics symposium. Sydney University Press, Brisbane, pp 14–15
van Ooijen JW (2006) JoinMap® 4.0: software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Weaver R, Helms C, Mishra SK, Donis-Keller H (1992) Software for analysis and manipulation of genetic linkage data. Am J Hum Genet 50(6):1267–1274
Wu Y, Close TJ, Lonardi S (2008) On the accurate construction of consensus genetic maps. Comput Syst Bioinform Conf 7:285–296
Würschum T (2012) Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125(2):201–210
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178(1):539–551
Zhang X, Bai G, Bockus W, Ji X, Pan H (2012) Quantitative trait loci for fusarium head blight resistance in U.S. Hard winter wheat cultivar Heyne. Crop Sci 52(3):1187–1194
Acknowledgments
The authors acknowledge support from the US Wheat and Barley Scab Initiative under ARS Agreement No: 59-0200-3-005 to J.L.G.H. and by the South Dakota Agricultural Experimental Station.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eckard, J.T., Gonzalez-Hernandez, J.L., Chao, S. et al. Construction of dense linkage maps “on the fly” using early generation wheat breeding populations. Mol Breeding 34, 1281–1300 (2014). https://doi.org/10.1007/s11032-014-0116-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0116-1