Abstract
To date, many HDAC6 inhibitors have been identified and developed but none is clinically approved as of now. Through this study, we aim to obtain novel HDAC6 selective inhibitors and provide new insights into the detailed structural design of potential HDAC6 inhibitors. A HypoGen-based 3D QSAR HDAC6 pharmacophore was built and used as a query model to screen approximately 8 million ZINC database compounds. First, the ZINC Database was filtered using ADMET, followed by pharmacophore-based library screening. Using fit value and estimated activity cutoffs, a final set of 54 ZINC hits was obtained that were further investigated using molecular docking with the crystal structure of human histone deacetylase 6 catalytic domain 2 in complex with Trichostatin A (PDB ID: 5EDU). Through detailed in silico screening of the ZINC database, we shortlisted three hits as the lead molecules for designing novel HDAC6 inhibitors with better efficacy. Docking with 5EDU, followed by ADMET and TOPKAT analysis of modified ZINC hits provided 9 novel potential HDAC6 inhibitors that possess better docking scores and 2D interactions as compared to the control ZINC hit molecules. Finally, a 50 ns MD analysis run followed by Protein–Ligand Interaction Energy (PLIE) analysis of the top scored hits provided a novel molecule N1 that showed promisingly similar results to that of Ricolinostat (a known HDAC6 inhibitor). The comparable result of the designed hits to established HDAC6 inhibitors suggests that these compounds might prove to be successful HDAC6 inhibitors in future.
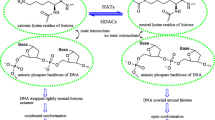
Graphical abstract
Designed novel hits that might act as good HDAC6 inhibitors derived from ZINC database using combined molecular docking and modeling approaches.









Similar content being viewed by others
Abbreviations
- –CDIE:
-
–CDOCKER Interaction Energy
- HDAC6i:
-
HDAC6 inhibitor
- 2D:
-
2 Dimensional
- TSA:
-
Trichostatin A
- Tub A:
-
Tubastatin A
- ZBG:
-
Zinc Binding Group
References
Li Y, Li Z, Zhu W-G (2019) Molecular mechanisms of epigenetic regulators as activatable targets in cancer theranostics. Curr Med Chem 26:1328–1350. https://doi.org/10.2174/0929867324666170921101947
Wang X-X, Wan R-Z, Liu Z-P (2018) Recent advances in the discovery of potent and selective HDAC6 inhibitors. Eur J Med Chem 143:1406–1418. https://doi.org/10.1016/j.ejmech.2017.10.040
Biancotto C, Frigè G, Minucci S (2010) Histone modification therapy of cancer. Adv Genet 70:341–386. https://doi.org/10.1016/B978-0-12-380866-0.60013-7
Ropero S, Esteller M (2007) The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 1:19–25. https://doi.org/10.1016/j.molonc.2007.01.001
Audia JE, Campbell RM (2016) Histone modifications and cancer. Cold Spring Harb Perspect Biol 8:a019521. https://doi.org/10.1101/cshperspect.a019521
Okugawa Y, Grady WM, Goel A (2015) Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149:1204-1225.e12. https://doi.org/10.1053/j.gastro.2015.07.011
Li T, Zhang C, Hassan S et al (2018) Histone deacetylase 6 in cancer. J Hematol Oncol 11:111. https://doi.org/10.1186/s13045-018-0654-9
Hao M, Song F, Du X et al (2015) Advances in targeted therapy for unresectable melanoma: New drugs and combinations. Cancer Lett 359:1–8. https://doi.org/10.1016/j.canlet.2014.12.050
Zhang X, Yuan Z, Zhang Y et al (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell 27:197–213. https://doi.org/10.1016/j.molcel.2007.05.033
Basu R, Wu S, Kopchick JJ (2017) Targeting growth hormone receptor in human melanoma cells attenuates tumor progression and epithelial mesenchymal transition via suppression of multiple oncogenic pathways. Oncotarget 8:21579–21598. https://doi.org/10.18632/oncotarget.15375
Zhang L, Sheng S, Qin C (2013) The role of HDAC6 in Alzheimer’s disease. J Alzheimers Dis 33:283–295. https://doi.org/10.3233/JAD-2012-120727
Selenica M-L, Benner L, Housley SB et al (2014) Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimers Res Ther 6:12. https://doi.org/10.1186/alzrt241
Du G, Liu X, Chen X et al (2010) Drosophila histone deacetylase 6 protects dopaminergic neurons against {alpha}-synuclein toxicity by promoting inclusion formation. Mol Biol Cell 21:2128–2137. https://doi.org/10.1091/mbc.e10-03-0200
McLendon PM, Ferguson BS, Osinska H et al (2014) Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci U S A 111:E5178-5186. https://doi.org/10.1073/pnas.1415589111
Dickinson M, Johnstone RW, Prince HM (2010) Histone deacetylase inhibitors: potential targets responsible for their anti-cancer effect. Invest New Drugs 28(Suppl 1):S3-20. https://doi.org/10.1007/s10637-010-9596-y
Butler KV, Kozikowski AP (2008) Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des 14:505–528. https://doi.org/10.2174/138161208783885353
Wang D-F, Helquist P, Wiech NL, Wiest O (2005) Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human Class I histone deacetylases. J Med Chem 48:6936–6947. https://doi.org/10.1021/jm0505011
Furumai R, Komatsu Y, Nishino N et al (2001) Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci U S A 98:87–92. https://doi.org/10.1073/pnas.011405598
Dallavalle S, Pisano C, Zunino F (2012) Development and therapeutic impact of HDAC6-selective inhibitors. Biochem Pharmacol 84:756–765. https://doi.org/10.1016/j.bcp.2012.06.014
Osko JD, Porter NJ, Narayana Reddy PA et al (2020) Exploring structural determinants of inhibitor affinity and selectivity in complexes with histone deacetylase 6. J Med Chem 63:295–308. https://doi.org/10.1021/acs.jmedchem.9b01540
Porter NJ, Mahendran A, Breslow R, Christianson DW (2017) Unusual zinc-binding mode of HDAC6-selective hydroxamate inhibitors. Proc Natl Acad Sci U S A 114:13459–13464. https://doi.org/10.1073/pnas.1718823114
Suzuki T (2009) Explorative study on isoform-selective histone deacetylase inhibitors. Chem Pharm Bull (Tokyo) 57:897–906. https://doi.org/10.1248/cpb.57.897
Sharma M, Jha P, Verma P, Chopra M (2019) Combined comparative molecular field analysis, comparative molecular similarity indices analysis, molecular docking and molecular dynamics studies of histone deacetylase 6 inhibitors. Chem Biol Drug Des 93:910–925. https://doi.org/10.1111/cbdd.13488
Liu J, Zhu Y, He Y et al (2020) Combined pharmacophore modeling, 3D-QSAR and docking studies to identify novel HDAC inhibitors using drug repurposing. J Biomol Struct Dyn 38:533–547. https://doi.org/10.1080/07391102.2019.1590241
Subramanian S, Bates SE, Wright JJ et al (2010) Clinical toxicities of histone deacetylase inhibitors. Pharmaceuticals (Basel, Switzerland). https://doi.org/10.3390/ph3092751
Auzmendi-Iriarte J, Saenz-Antoñanzas A, Mikelez-Alonso I et al (2020) Characterization of a new small-molecule inhibitor of HDAC6 in glioblastoma. Cell Death Dis 11:1–14. https://doi.org/10.1038/s41419-020-2586-x
Zhang Y, Kwon S, Yamaguchi T et al (2008) Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. https://doi.org/10.1128/MCB.01154-06
Senger J, Melesina J, Marek M et al (2016) Synthesis and biological investigation of oxazole hydroxamates as highly selective histone deacetylase 6 (HDAC6) inhibitors. J Med Chem 59:1545–1555. https://doi.org/10.1021/acs.jmedchem.5b01493
Uba AI, Yelekçi K (2018) Pharmacophore-based virtual screening for identification of potential selective inhibitors of human histone deacetylase 6. Comput Biol Chem 77:318–330. https://doi.org/10.1016/j.compbiolchem.2018.10.016
Lee J-H, Mahendran A, Yao Y et al (2013) Development of a histone deacetylase 6 inhibitor and its biological effects. Proc Natl Acad Sci U S A 110:15704–15709. https://doi.org/10.1073/pnas.1313893110
Wang Y, Yang L, Hou J et al (2019) Hierarchical virtual screening of the dual MMP-2/HDAC-6 inhibitors from natural products based on pharmacophore models and molecular docking. J Biomol Struct Dyn 37:649–670. https://doi.org/10.1080/07391102.2018.1434833
Zeb A, Park C, Rampogu S et al (2019) Structure-based drug designing recommends HDAC6 inhibitors to attenuate microtubule-associated tau-pathogenesis. ACS Chem Neurosci 10:1326–1335. https://doi.org/10.1021/acschemneuro.8b00405
Song H, Niu X, Quan J et al (2020) Discovery of specific HDAC6 inhibitor with anti-metastatic effects in pancreatic cancer cells through virtual screening and biological evaluation. Bioorg Chem 97:103679. https://doi.org/10.1016/j.bioorg.2020.103679
Yan G, Li D, Zhong X et al (2021) Identification of HDAC6 selective inhibitors: pharmacophore based virtual screening, molecular docking and molecular dynamics simulation. J Biomol Struct Dyn 39:1928–1939. https://doi.org/10.1080/07391102.2020.1743760
Sterling T, Irwin JJ (2022) ZINC 15—ligand discovery for everyone. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.5b00559
Van Der Spoel D, Lindahl E, Hess B et al (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Eklund EA, Platanias LC (2011) Inhibition of histone deacetylase 6 as a therapeutic strategy for acute lymphocytic leukemia. Leuk Lymphoma 52:1421–1422. https://doi.org/10.3109/10428194.2011.577259
Kalin JH, Butler KV, Akimova T et al (2012) Second-generation histone deacetylase 6 inhibitors enhance the Immunosuppressive effects of Foxp3+ T-regulatory cells. J Med Chem 55:639–651. https://doi.org/10.1021/jm200773h
Gupta PK, Reid RC, Liu L et al (2010) Inhibitors selective for HDAC6 in enzymes and cells. Bioorg Med Chem Lett 20:7067–7070. https://doi.org/10.1016/j.bmcl.2010.09.100
Jones P, Bottomley MJ, Carfí A et al (2008) 2-Trifluoroacetylthiophenes, a novel series of potent and selective class II histone deacetylase inhibitors. Bioorg Med Chem Lett 18:3456–3461. https://doi.org/10.1016/j.bmcl.2008.02.026
Kozikowski AP, Tapadar S, Luchini DN et al (2008) Use of the nitrile oxide cycloaddition (NOC) reaction for molecular probe generation: a new class of enzyme selective histone deacetylase inhibitors (HDACIs) showing picomolar activity at HDAC6. J Med Chem 51:4370–4373. https://doi.org/10.1021/jm8002894
Itoh Y, Suzuki T, Kouketsu A et al (2007) Design, synthesis, structure−selectivity relationship, and effect on human cancer cells of a novel series of histone deacetylase 6-selective inhibitors. J Med Chem 50:5425–5438. https://doi.org/10.1021/jm7009217
Scarpelli R, Di Marco A, Ferrigno F et al (2008) Studies of the metabolic stability in cells of 5-(trifluoroacetyl)thiophene-2-carboxamides and identification of more stable class II histone deacetylase (HDAC) inhibitors. Bioorg Med Chem Lett 18:6078–6082. https://doi.org/10.1016/j.bmcl.2008.10.041
Ontoria JM, Altamura S, Di Marco A et al (2009) Identification of novel, selective, and stable inhibitors of class ii histone deacetylases. Validation studies of the inhibition of the enzymatic activity of HDAC4 by small molecules as a novel approach for cancer therapy. J Med Chem 52:6782–6789. https://doi.org/10.1021/jm900555u
Chen Y, Lopez-Sanchez M, Savoy DN et al (2008) A series of potent and selective, triazolylphenyl-based histone deacetylases inhibitors with activity against pancreatic cancer cells and Plasmodium falciparum. J Med Chem 51:3437–3448. https://doi.org/10.1021/jm701606b
Kozikowski AP, Chen Y, Gaysin AM et al (2008) Chemistry, biology, and QSAR studies of substituted biaryl hydroxamates and mercaptoacetamides as HDAC inhibitors—nanomolar-potency inhibitors of pancreatic cancer cell growth. ChemMedChem 3:487–501. https://doi.org/10.1002/cmdc.200700314
Inks ES, Josey BJ, Jesinkey SR, Chou CJ (2012) A novel class of small molecule inhibitors of HDAC6. ACS Chem Biol 7:331–339. https://doi.org/10.1021/cb200134p
Auzzas L, Larsson A, Matera R et al (2010) Non-natural macrocyclic inhibitors of histone deacetylases: design, synthesis, and activity. J Med Chem 53:8387–8399. https://doi.org/10.1021/jm101092u
Smil DV, Manku S, Chantigny YA et al (2009) Novel HDAC6 isoform selective chiral small molecule histone deacetylase inhibitors. Bioorg Med Chem Lett 19:688–692. https://doi.org/10.1016/j.bmcl.2008.12.045
Dallavalle S, Cincinelli R, Nannei R et al (2009) Design, synthesis, and evaluation of biphenyl-4-yl-acrylohydroxamic acid derivatives as histone deacetylase (HDAC) inhibitors. Eur J Med Chem 44:1900–1912. https://doi.org/10.1016/j.ejmech.2008.11.005
Olsen CA, Ghadiri MR (2009) Discovery of potent and selective histone deacetylase inhibitors via focused combinatorial libraries of cyclic α3β-tetrapeptides. J Med Chem 52:7836–7846. https://doi.org/10.1021/jm900850t
Liu T, Lin Y, Wen X et al (2007) BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35:D198–D201. https://doi.org/10.1093/nar/gkl999
Bertrand P (2010) Inside HDAC with HDAC inhibitors. Eur J Med Chem 45:2095–2116. https://doi.org/10.1016/j.ejmech.2010.02.030
Dai Y, Guo Y, Curtin ML et al (2003) A novel series of histone deacetylase inhibitors incorporating hetero aromatic ring systems as connection units. Bioorg Med Chem Lett 13:3817–3820. https://doi.org/10.1016/j.bmcl.2003.07.012
Wang H, Lim Z-Y, Zhou Y et al (2010) Acylurea connected straight chain hydroxamates as novel histone deacetylase inhibitors: synthesis, SAR, and in vivo antitumor activity. Bioorg Med Chem Lett 20:3314–3321. https://doi.org/10.1016/j.bmcl.2010.04.041
Lavoie R, Bouchain G, Frechette S et al (2001) Design and synthesis of a novel class of histone deacetylase inhibitors. Bioorg Med Chem Lett 11:2847–2850. https://doi.org/10.1016/S0960-894X(01)00552-2
Nishino N, Yoshikawa D, Watanabe LA et al (2004) Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg Med Chem Lett 14:2427–2431. https://doi.org/10.1016/j.bmcl.2004.03.018
Mai A, Massa S, Cerbara I et al (2004) 3-(4-Aroyl-1-methyl-1H-2-pyrrolyl)-N-hydroxy-2-propenamides as a new class of synthetic histone deacetylase inhibitors. 2. Effect of pyrrole-C2 and/or -C4 substitutions on biological activity. J Med Chem 47:1098–1109. https://doi.org/10.1021/jm030990+
Chen Y, Jiang Y-J, Zhou J-W et al (2008) Identification of ligand features essential for HDACs inhibitors by pharmacophore modeling. J Mol Graph Model 26:1160–1168. https://doi.org/10.1016/j.jmgm.2007.10.007
de Ruijter AJM, van Gennip AH, Caron HN et al (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370:737–749. https://doi.org/10.1042/BJ20021321
Finnin MS, Donigian JR, Cohen A et al (1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188–193. https://doi.org/10.1038/43710
Hu E, Dul E, Sung C-M et al (2003) Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther 307:720–728. https://doi.org/10.1124/jpet.103.055541
Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1:287–299. https://doi.org/10.1038/nrd772
Vadivelan S, Sinha BN, Rambabu G et al (2008) Pharmacophore modeling and virtual screening studies to design some potential histone deacetylase inhibitors as new leads. J Mol Graph Model 26:935–946. https://doi.org/10.1016/j.jmgm.2007.07.002
Tang H, Wang XS, Huang X-P et al (2009) Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J Chem Inf Model 49:461–476. https://doi.org/10.1021/ci800366f
Yu L, Liu F, Chen Y, You Q (2009) Pharmacophore identification of hydroxamate HDAC 1 inhibitors. Chin J Chem 27:557–564. https://doi.org/10.1002/cjoc.200990091
Chopra M, Gupta R, Gupta S, Saluja D (2008) Molecular modeling study on chemically diverse series of cyclooxygenase-2 selective inhibitors: generation of predictive pharmacophore model using catalyst. J Mol Model 14:1087–1099. https://doi.org/10.1007/s00894-008-0350-8
Chopra M, Mishra AK (2005) Ligand-based molecular modeling study on a chemically diverse series of cholecystokinin-B/gastrin receptor antagonists: generation of predictive model. J Chem Inf Model 45:1934–1942. https://doi.org/10.1021/ci050257m
Verma P, Dalal K, Chopra M (2016) Pharmacophore development and screening for discovery of potential inhibitors of ADAMTS-4 for osteoarthritis therapy. J Mol Model 22:178. https://doi.org/10.1007/s00894-016-3035-8
Dong X, Yan J, Du L et al (2012) Pharmacophore identification, docking and “in silico” screening for novel CDK1 inhibitors. J Mol Graph Model 37:77–86. https://doi.org/10.1016/j.jmgm.2012.04.003
Bickerton GR, Paolini GV, Besnard J et al (2012) Quantifying the chemical beauty of drugs. Nature Chem 4:90–98. https://doi.org/10.1038/nchem.1243
Pal S, Kumar V, Kundu B et al (2019) Ligand-based pharmacophore modeling, virtual screening and molecular docking studies for discovery of potential topoisomerase I inhibitors. Comput Struct Biotechnol J 17:291–310. https://doi.org/10.1016/j.csbj.2019.02.006
Bank RPD RCSB PDB: Homepage. https://www.rcsb.org/. Accessed 19 Apr 2022
Hai Y, Christianson DW (2016) Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol 12:741–747. https://doi.org/10.1038/nchembio.2134
Osko JD, Christianson DW (2020) Structural determinants of affinity and selectivity in the binding of inhibitors to histone deacetylase 6. Bioorg Med Chem Lett 30:127023. https://doi.org/10.1016/j.bmcl.2020.127023
Baby ST, Sharma S, Enaganti S, Cherian PR (2016) Molecular docking and pharmacophore studies of heterocyclic compounds as Heat shock protein 90 (Hsp90) Inhibitors. Bioinformation 12:149–155. https://doi.org/10.6026/97320630012149
Yuan G, Li T, Hu W (2019) A conjugate gradient algorithm and its application in large-scale optimization problems and image restoration. J Inequal Appl 2019:247. https://doi.org/10.1186/s13660-019-2192-6
Abdel-Hamid MK, McCluskey A (2014) In silico docking, molecular dynamics and binding energy insights into the bolinaquinone-clathrin terminal domain binding site. Molecules 19:6609–6622. https://doi.org/10.3390/molecules19056609
Wu G, Robertson DH, Brooks CL III, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER—A CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562. https://doi.org/10.1002/jcc.10306
Ding X, Wu Y, Wang Y et al (2020) Accelerated CDOCKER with GPUs, parallel simulated annealing, and fast Fourier transforms. J Chem Theory Comput 16:3910–3919. https://doi.org/10.1021/acs.jctc.0c00145
Li X, Li Y, Cheng T et al (2010) Evaluation of the performance of four molecular docking programs on a diverse set of protein–ligand complexes. J Comput Chem 31:2109–2125. https://doi.org/10.1002/jcc.21498
Li Y, Su M, Liu Z et al (2018) Assessing protein–ligand interaction scoring functions with the CASF-2013 benchmark. Nat Protoc 13:666–680. https://doi.org/10.1038/nprot.2017.114
Krammer A, Kirchhoff PD, Jiang X et al (2005) LigScore: a novel scoring function for predicting binding affinities. J Mol Graph Model 23:395–407. https://doi.org/10.1016/j.jmgm.2004.11.007
Chen P-Y, Tsai C-T, Ou C-Y et al (2012) Computational analysis of novel drugs designed for use as acetylcholinesterase inhibitors and histamine H3 receptor antagonists for Alzheimer’s disease by docking, scoring and de novo evolution. Mol Med Rep 5:1043–1048. https://doi.org/10.3892/mmr.2012.757
Zhu Y, Han Y, Ma Y, Yang P (2018) ADME/toxicity prediction and antitumor activity of novel nitrogenous heterocyclic compounds designed by computer targeting of alkylglycerone phosphate synthase. Oncol Lett 16:1431–1438. https://doi.org/10.3892/ol.2018.8873
Schüttelkopf AW, van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Cortopassi WA, Penna-Coutinho J, Aguiar ACC et al (2014) Theoretical and experimental studies of new modified isoflavonoids as potential inhibitors of topoisomerase I from Plasmodium falciparum. PLoS ONE 9:e91191. https://doi.org/10.1371/journal.pone.0091191
Bepari AK, Reza HM (2021) Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation. PeerJ 9:e11261. https://doi.org/10.7717/peerj.11261
Cosenza M, Civallero M, Marcheselli L et al (2017) Ricolinostat, a selective HDAC6 inhibitor, shows anti-lymphoma cell activity alone and in combination with bendamustine. Apoptosis 22:827–840. https://doi.org/10.1007/s10495-017-1364-4
Raje NS, Bensinger W, Cole CE et al (2015) Ricolinostat (ACY-1215), the first selective HDAC6 inhibitor, combines safely with pomalidomide and dexamethasone and shows promosing early results in relapsed-and-refractory myeloma (ACE-MM-102 study). Blood 126:4228. https://doi.org/10.1182/blood.V126.23.4228.4228
Cheng F, Laino AS, Wang H et al (2016) In vitro and in vivo anti-melanoma activity of ricolinostat, a selective HDAC6 inhibitor with immunomodulatory properties. JCO 34:e21075–e21075. https://doi.org/10.1200/JCO.2016.34.15_suppl.e21075
Cao J, Lv W, Wang L et al (2018) Ricolinostat (ACY-1215) suppresses proliferation and promotes apoptosis in esophageal squamous cell carcinoma via miR-30d/PI3K/AKT/mTOR and ERK pathways. Cell Death Dis 9:1–11. https://doi.org/10.1038/s41419-018-0788-2
Li L, Liu F, Huang W et al (2019) Ricolinostat (ACY-1215) inhibits VEGF expression via PI3K/AKT pathway and promotes apoptosis in osteoarthritic osteoblasts. Biomed Pharmacother 118:109357. https://doi.org/10.1016/j.biopha.2019.109357
Acknowledgements
Priya Poonia would like to acknowledge ICMR for providing her SRF fellowship. Prakash Jha would like to thank the Department of Science and Technology, Government of India for awarding him DST-INSPIRE fellowship (Grant Number: DST/INSPIRE/03/2016/000026).
Funding
The authors would like to thank Dr. B.R. Ambedkar Center for Biomedical Research (ACBR), University of Delhi for providing the lab space and instrument facilities; DBT-BIC facility (BT/PR40153/137/8/2021) at ACBR from Department of Biotechnology, Government of India for providing software and hardware facility.
Author information
Authors and Affiliations
Contributions
PP carried out the molecular docking and molecular modeling studies. MS carried out pharmacophore development and validation. MC conceived, designed, and coordinated the study. PJ carried out MD simulations, and compiled the methodology and results of the same. PP wrote the whole manuscript, and prepared the figures and tables. MC reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there is no conflict of interest.
Ethical approval
There was no human or animal experiment conducted for this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11030_2022_10540_MOESM1_ESM.docx
Supplementary file1 Charts for training set and test set compounds used to build HDAC6 and HDAC1/2 pharmacophore. Result of pharmacophore validation and molecular docking results of ZINC hits (DOCX 2676 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Poonia, P., Sharma, M., Jha, P. et al. Pharmacophore-based virtual screening of ZINC database, molecular modeling and designing new derivatives as potential HDAC6 inhibitors. Mol Divers 27, 2053–2071 (2023). https://doi.org/10.1007/s11030-022-10540-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10540-3