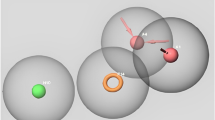
Abstract
Diabetes mellitus (DM) is one of the major health problems worldwide. WHO have estimated that 439 million people may have DM by the year 2030. Several classes of drugs such as sulfonylureas, meglitinides, thiazolidinediones etc. are available to manage this disease, however, there is no cure for this disease. Salt inducible kinase 2 (SIK2) is expressed several folds in adipose tissue than in normal tissues and thus SIK2 is one of the attractive targets for DM treatment. SIK2 inhibition improves glucose homeostasis. Several analogues have been reported and experimentally proven against SIK for DM treatment. But, identifying potential SIK2 inhibitors with improved efficacy and good pharmacokinetic profiles will be helpful for the effective treatment of DM. The objective of the present study is to identify selective SIK2 inhibitors with good pharmacokinetic profiles. Due to the unavailability of SIK2 structure, the modeled structure of SIK2 will be an important to understand the atomic level of SIK2 inhibitors in the binding site pocket. In this study, different molecular modeling studies such as Homology Modeling, Molecular Docking, Pharmacophore-based virtual screening, MD simulations, Density Functional Theory calculations and WaterMap analysis were performed to identify potential SIK2 inhibitors. Five molecules from different databases such as Binding_4067, TosLab_837067, NCI_349155, Life chemicals_ F2565-0113, Enamine_7623111186 molecules were identified as possible SIK2 inhibitors.
Graphical Abstract















Similar content being viewed by others
Abbreviations
- ADME:
-
Absorption distribution metabolism excretion
- CAMK:
-
Calcium/calmodulin kinase
- CREB:
-
CAMP-response element binding protein
- CRTCs:
-
CREB-regulated transcriptional coactivators
- CADD:
-
Computer-aided drug design
- DM:
-
Diabetes mellitus
- DFT:
-
Density functional theory
- DOPE:
-
Discrete optimized protein energy
- GCMC:
-
Grand canonical Monte Carlo
- GCAP:
-
Grand canonical alchemical perturbation
- GPCR:
-
G protein-coupled receptor
- GROMACS:
-
GROningen MAchine for chemical simulations
- HDACs:
-
Histone deacetylases
- HTVS:
-
High-throughput virtual screening
- HOMO:
-
Highest occupied molecular orbital
- IST:
-
Inhomogeneous solvation theory
- LKB1:
-
Liver kinase B1
- LUMO:
-
Lowest unoccupied molecular orbital
- MDS:
-
Molecular dynamic simulation
- MDCK:
-
Madin-Darby canine kidney
- MM-PBSA:
-
Molecular mechanics Poisson-Boltzmann surface area
- NVT:
-
Constant temperature, constant volume
- NPT:
-
Constant temperature, constant pressure
- OPLS3e:
-
Optimized potentials for liquid simulations
- PDB:
-
Protein data bank
- ProSA:
-
Protein structural analysis
- PBF:
-
Poisson Boltzmann
- SGLT2:
-
Sodium-glucose co-transporter-2 finite
- SP:
-
Standard precision
- SPC:
-
Simple point charge
- STM:
-
Single trajectory method
- T2DM:
-
Type 2 Diabetes Mellitus
- WHO:
-
World Health Organization
- SIK2:
-
Salt inducible kinase 2
- WM:
-
WaterMap
- XP:
-
Extra precision
References
Stumvol M, Goldstein BJ, Van Haeften TW (2005) Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365(9467):1333–1346. https://doi.org/10.1016/S0140-6736(05)61032-X
Lee PG, Halter JB (2017) The Pathophysiology of hyperglycemia in older adults: clinical considerations. Diabetes Care 40(4):444–452. https://doi.org/10.2337/dc16-1732
Ryu TY, Park J, Scherer PE (2014) Hyperglycemia as a risk factor for cancer progression. Diabetes Metab J 38(5):330–336. https://doi.org/10.4093/dmj.2014.38.5.330
Tachkov K, Mitov K, Koleva Y, Mitkova Z, Kamusheva M, Dimitrova M, Petkova V, Savova A, Doneva M, Tcarukciev D, Valov V, Angelova G, Manova M, Petrova G (2020) Life expectancy and survival analysis of patients with diabetes compared to the non diabetic population in Bulgaria. PLOS ONE 15(5):e0232815. https://doi.org/10.1371/journal.pone.0232815
Sarwar N, Gao P, Seshasai GR, Gobin R, Kaptoge S, Di Angelantonio E, Ingelsson E, Lawlor DA, Selvin E, Stampfer M, Stehouwer CD, Lewington S, Pennells L, Thompson A, Sattar N, White IR, Ray KK, Danesh J (2010) Diabetes mellitus, fasting blood glucose concentration and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet 375(9733):2215–22. https://doi.org/10.1016/S0140-6736(10)60484-9
Divers J, Mayer-Davis EJ, Lawrence JM, Isom S, Dabelea D, Dolan L, Imperatore G, Marcovina S, Pettitt DJ, Pihoker C, Hamman RF, Saydah S, Wagenknecht LE (2020) Trends in incidence of type 1 and type 2 diabetes among youths—selected counties and Indian reservations, United States, 2002–2015. MMWR Morb Mortal Wkly Rep 69(6):161–165. https://doi.org/10.15585/mmwr.mm6906a3
Padhi S, Nayak AK, Behera A (2020) Type II diabetes mellitus: a review on recent drug based therapeutics. Biomed Pharmacother 131:110708. https://doi.org/10.1016/j.biopha.2020.110708
Cahn Avivit, Cefalu William T (2016) Clinical considerations for use of initial combination therapy in type 2 diabetes. Diabetes Care 39(Supplement_2):S137–S145. https://doi.org/10.2337/dcS15-3007
Feingold KR (2000) Oral and Injectable (Non-insulin) Pharmacological Agents for Type 2 Diabetes, MD Text. com Inc South Dartmouth (MA)
Sun Z, Jiang Q, Li J, Guo J (2020) The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis. Signal Transduct Target Ther 5(1):150. https://doi.org/10.1038/s41392-020-00265-w
Park J, Yoon YS, Han HS, Kim YH, Ogawa Y, Park KG, Lee CH, Kim ST, Koo SH (2014) SIK2 Is critical in the regulation of lipid homeostasis and adipogenesis in Vivo. Diabetes 63:3659–3673. https://doi.org/10.2337/db13-1423
Henriksson E, Sall J, Gormand A, Wasserstrom S, Morrice NA, Fritzen AM, Foretz M, Campbell DG, Sakamoto K, Ekelund M, Degerman E, Stenkula KG, Goransson O (2015) SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. J Cell Sci 128:472–486. https://doi.org/10.1242/jcs.153932
Wyss PC, Gerber P, Hartman PG, Hubschwerlen C, Locher H, Marty HP, Stahl M (2003) Novel dihydrofolate reductase inhibitors. Structure-based versus diversity-based library design and high-throughput synthesis and screening. J Med Chem 46(12):2304–2312. https://doi.org/10.1021/jm020495y
Alessi DR, Sakamoto K, Bayascas JR (2006) LKB1-dependent signaling pathways. Annu Rev Biochem 75(1):137–163. https://doi.org/10.1146/annurev.biochem.75.103004.142702
Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280(32):29060–29066. https://doi.org/10.1074/jbc.M503824200
Sakamoto K, Bultot L, Göransson O (2018) The salt-inducible kinases: emerging metabolic regulators. Trends Endocrinol Metab 29(12):827–840. https://doi.org/10.1016/j.tem.2018.09.007
Säll J, Pettersson AML, Björk C, Henriksson E, Wasserstrom S, Linder W, Zhou Y, Hansson O, Andersson DP, Ekelund M, Degerman E, Stenkula KG, Laurencikiene J, Göransson O (2016) Salt-inducible kinase 2 and -3 are downregulated in adipose tissue from obese or insulin-resistant individuals: implications for insulin signalling and glucose uptake in human adipocytes. Diabetologia 60(2):314–323. https://doi.org/10.1007/s00125-016-4141-y
Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M (2007) SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med 13(5):597–603. https://doi.org/10.1038/nm1573
Walkinshaw DR, Weist R, Kim GW, You L, Xiao L, Nie J, Li CS, Zhao S, Minghong Xu, Yang XJ (2013) The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J Biol Chem 288(13):9345–9362. https://doi.org/10.1074/jbc.M113.456996
Patel K, Foretz M, Marion A, Campbell DG, Gourlay R, Boudaba N, Tournier E, Titchenell P, Peggie M, Deak M, Wan M, Kaestner KH, Göransson O, Viollet B, Gray NS, Birnbaum MJ, Sutherland C, Sakamoto K (2014) The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nat Commun. https://doi.org/10.1038/ncomms5535
Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-González E (2014) Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci 5:190. https://doi.org/10.3389/fpls.2014.00190
Hedbacker K, Carlson M (2008) SNF1/AMPK pathways in yeast. Front Biosci 13:2408–2420. https://doi.org/10.2741/2854
Sonntag T, Vaughan JM, Montminy M (2018) 14–3–3 proteins mediate inhibitory effects of cAMP on salt-inducible kinases (SIKs). FEBS J 285(3):467–480. https://doi.org/10.1111/febs.14351
Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet Brian K (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1b. J Med Chem 45(11):2213–2221. https://doi.org/10.1021/jm010548w
Kick EK, Roe DC, Skillman AG, Liu G, Ewing TJ, Sun Y, Kuntz ID, Ellman JA (1997) Structure-based design and combinatorial chemistry yield low nanomolar inhibitors of cathepsin D. Chem Biol 4:297–307. https://doi.org/10.1016/s1074-5521(97)90073-9
Oshiro C, Bradley EK, Eksterowicz J, Evensen E, Lamb ML, Lanctot JK, Putta S, Stanton R, Grootenhuis PD (2004) Performance of 3d-database molecular docking studies into homology models. J Med Chem 47(3):764–767. https://doi.org/10.1021/jm0300781
Paiva AM, Vanderwall DE, Blanchard JS, Kozarich JW, Williamson JM, Kelly TM (2001) Inhibitors of dihydrodipicolinate reductase, a key enzyme of the diaminopimelate pathway of Mycobacterium tuberculosis. Biochimica et Biophysica Acta 1545(1–2):67–77. https://doi.org/10.1016/S0167-4838(00)00262-4
Cavasotto CN, Phatak SS (2009) Homology modeling in drug discovery: current trends and applications. Drug Discov Today 14:676–683. https://doi.org/10.1016/j.drudis.2009.04.006
UniProt Consortium, Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R et al (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49(D1):D480-89. https://doi.org/10.1093/nar/gkaa1100
Boratyn GM, Thierry-Mieg J, Thierry-Mieg D, Busby B, Madden TL (2019) Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics. https://doi.org/10.1186/s12859-019-2996-x
Shi J, Blundell TL, Mizuguchi K (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Molecular Biol 310(1):243–257. https://doi.org/10.1006/jmbi.2001.4762
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinform. https://doi.org/10.1002/cpbi.3
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereo chemical quality of protein structures. J Appl Crystallogr 26(2):283–291. https://doi.org/10.1107/S0021889892009944
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(Web Server):W407–W410. https://doi.org/10.1093/nar/gkm290
Schrödinger Release 2021–4: (2021) Protein Preparation Wizard; Epik Schrödinger LLC New York NY 2021, Impact Schrödinger, LLC, New York, NY; Prime, Schrödinger, LLC, New York, NY.
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetic and properties of organic liquids. J Am Chem 118:11225–11236. https://doi.org/10.1021/ja9621760
Hosseini M, Chen W, Xiao D, Wang C (2021) Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs. Precis Clin Med 4(1):1–16. https://doi.org/10.1093/pcmedi/pbab001
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided-Drug Design 7(2):146–157. https://doi.org/10.2174/157340911795677602
Sundberg TB, Liang Y, Huixian W, Choi HG, Kim ND, Sim T, Johannessen L, Petrone A, Khor B, Graham DB, Latorre IJ, Phillips AJ, Schreiber SL, Perez J, Shamji AF, Gray NS, Xavier RJ (2016) Development of chemical probes for investigation of salt-inducible kinase function in vivo. ACS Chem Biol 11(8):2105–2111. https://doi.org/10.1021/acschembio.6b00217
Clark K, MacKenzie KF, Petkevicius K, Kristariyanto Y, Zhang J, Choi HG, Peggie M, Plater L, Pedrioli PGA, McIver Ed, Gray NS, Simon J, Arthur C, Cohen P (2012) Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci 109(42):16986–16991. https://doi.org/10.1073/pnas.1215450109
Mingsong S, Lun W, Penghui L, Jiang L, Lijuan C, Dingguo X (2021) Dasatinib-SIK2 binding elucidated by homology modeling molecular docking and dynamics simulations. ACS Omega 6:11025–11038. https://doi.org/10.1021/acsomega.1c00947
Roberta T, Marcel R, Monika R, Lena MB, Thales K, Andreas CJ, Benedict-Tilman B, Ismahan A, Thomas H, Antti P, Klaus S, Mourad S, Stefan K (2021) Structure-based design of selective salt-inducible kinase inhibitors. J Med Chem 64:8142–8160. https://doi.org/10.1101/2021.04.08.439011
Shi M, Zhao M, Wang L, Liu K, Li P, Liu J, Xu D (2021) Exploring the stability of inhibitor binding to SIK2 using molecular dynamics simulation and binding free energy calculation. Phys Chem Chem Phys 23:13216–13227. https://doi.org/10.1039/D1CP00717C
Mills Nancy (2006) ChemDraw Ultra 10.0 CambridgeSoft, 100 CambridgePark drive, Cambridge, MA 02140. www.cambridgesoft.com. Commercial Price: $1910 for download, $2150 for CD-ROM; academic price: $710 for download, $800 for CD-ROM. J Am Chem Soc 128(41):13649–13650
Schrödinger Release 2020 (2020) LigPrep. Schrödinger LLC, New York
Roos K, Chuanjie W, Damm W, Reboul M, Stevenson JM, Chao L, Dahlgren MK, Mondal S, Chen W, Wang L, Abel R, Friesner RA, Harder ED (2019) OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput 15(3):1863–1874. https://doi.org/10.1021/acs.jctc.8b01026
Schrödinger Release 2020 (2020) Sitemap Schrödinger LLC, New York
Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49(2):377–389. https://doi.org/10.1021/ci800324m
Schrödinger Release 2020 (2020) Glide Schrödinger LLC, New York
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J Med Chem 47(7):1739–1749. https://doi.org/10.1021/jm0306430
Jayaprakash P, Biswal J, Kanagarajan S, Prabhu D, Gogoi P, Prasad Kanaujia S, Jeyakanthan J (2019) Design of novel PhMTNA inhibitors, targeting neurological disorder through homology modeling, molecular docking, and dynamics approaches. J Recept Signal Transduct 39(1):28–38. https://doi.org/10.1080/10799893.2019.1567786
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 9:6177–6196. https://doi.org/10.1021/jm051256o
Dixon SL, Smondyrev AM, Rao SN (2006) ö PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67:370–372. https://doi.org/10.1111/j.1747-0285.200600384.x
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA (2006) ö PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D Database screening. 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. https://doi.org/10.1007/s10822-006-9087-6
Biswal J, Jayaprakash P, Kumar RS, Venkatraman G, Poopandi S, Rangasamy R, Jeyaraman J (2019) Identification of Pak1 inhibitors using water thermodynamic analysis. J Biomol Struc Dyn 38(1):13–31. https://doi.org/10.1080/07391102.2019.1567393
Lionta E, Spyrou G, Vassilatis D, Cournia Z (2014) Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem 14(16):1923–1938. https://doi.org/10.2174/1568026614666140929124445
Moitessier N, Englebienne P, Lee DJ, Corbeil CR (2008) Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br J Pharmacol 153:S7–S26. https://doi.org/10.1038/sj.bjp.0707515
Rajamanikandan S, Jeyaraman J, Srinivasan P (2017) Discovery of potent inhibitors targeting Vibrio harveyi LuxR through shape and e-pharmacophore based virtual screening and its biological evaluation. Microb Pathog 103:40–56. https://doi.org/10.1016/j.micpath.2016.12.003
Choubey SK, Nachiappan M, Richard M, Chitra JP, Jeyaraman J (2020) Structural and functional insights of STAT2-NS5 interaction for the identification of NS5 antagonist-an approach for restoring interferon signaling. Comput Biol Chem 88:107332. https://doi.org/10.1016/j.compbiolchem.2020.107332
Choubey SK, Prabhu D, Nachiappan M, Biswal J, Jeyaraman J (2017) Molecular modeling, dynamics studies and density functional theory approaches to identify potential inhibitors of SIRT4 protein from Homo sapiens: a novel target for the treatment of type 2 diabetes. J Biomol Struct Dyn 35:3316–3329. https://doi.org/10.1080/07391102.2016.1254117
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7(2):146–57. https://doi.org/10.2174/157340911795677602
Acharya R, Chacko S, Bose P, Lapenna A, Prasad SP (2019) Structure based multitargeted molecular docking analysis of selected furanocoumarins against breast cancer. Sci rep 9:15743. https://doi.org/10.1038/s41598-019-52162-0
Schrödinger Release 2021–2 (2021) QikProp. Schrödinger LLC, New York
Prajisha J, Biswal J, Jeyakanthan J (2022) Discovery of potent Camkk1 kinase inhibitors through e-pharmacophore and molecular screening approaches. J Biomol Struct Dyn 40(6):2740–2756. https://doi.org/10.1080/07391102.2020.1842805
Schrödinger Release 2020 (2020) Watermap. Schrödinger LLC, New York
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136:B864–B871. https://doi.org/10.1103/PhysRev.136.B864
Bodnarchuk MS, Packer MJ, Haywood A (2020) Utilizing grand canonical Monte Carlo methods in drug discovery. ACS Med Chem Lett 11(1):77–82. https://doi.org/10.1021/acsmedchemlett.9b00499
Robinson DD, Sherman W, Farid R (2010) Understanding kinase selectivity through energetic analysis of binding site waters. Chem Med Chem 65(4):618–27. https://doi.org/10.1002/cmdc.200900501
Abel R, Young T, Farid R, Berne BJ, Friesner RA (2008) The role of the active site solvent in the thermodynamics of factor Xaligand binding. J Am Chem Soc 130:2817–2831. https://doi.org/10.1021/ja0771033
Mondal J, Friesner RA, Berne BJ (2014) Role of desolvation in thermodynamics and kinetics of ligand binding to a kinase. J Chem Theory Comput 10:5696–5705. https://doi.org/10.1021/ct500584n
Biswal J, Jayaprakash P, Rayala SK, Venkatraman G, Poopandi S, Rangaswamy R, Jeyaraman J (2021) Water mapping and scoring approaches to predict the role of hydration sites in binding affinity of PAK1 inhibitors. Comb Chem High Throughput Screen 25(4):660–676. https://doi.org/10.2174/1386207324666210308110646
Schrödinger Release 2021–3 (2021) Jaguar. Schrödinger LLC, New York
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Account 120:215–241. https://doi.org/10.1007/s00214-007-0310-x
Bochevarov AD, Harder E, Hughes TF, Greenwood JR, Braden DA, Philipp DM, Rinaldo D, Halls MD, Zhang J, Friesner RA (2013) Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem 113(18):2110–2142. https://doi.org/10.1002/qua.24481
Levy M (1979) Universal variational functionals of electron densities, first-order density matrices, and natural spin-orbitals and solution of the v-representability problem. Proc Natl Acad Sci 76(12):6062–6065. https://doi.org/10.1073/pnas.76.12.6062
Choubey SK, Jeyaraman J (2016) A mechanistic approach to explore novel HDAC1 inhibitor using pharmacophore modeling, 3D- QSAR analysis, molecular docking, density functional and molecular dynamics simulation study. J Mol Graph Model 70:54–69. https://doi.org/10.1016/j.jmgm.2016.09.008
Kohn W, Becke AD, Parr RG (1996) Density functional theory of electronic structure. J Phys Chem 100(31):12974–12980. https://doi.org/10.1021/jp960669l
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-e
Mai W, Li P, Bao H, Li X, Jiang L, Hu J, Werner DH (2019) Prism-based DGTD with a simplified periodic boundary condition to analyze FSS with D2n symmetry in a rectangular array under normal incidence. IEEE Antennas Wirel Propag Lett 18(4):771–75. https://doi.org/10.1109/LAWP.2019.2902340
Vander Sluis A, Van der Vorst HA (1986) The rate of convergence of conjugate gradients. Numer Math 48(5):543–560. https://doi.org/10.1007/BF01389450
Berendsen HJC, Postma JPM, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. https://doi.org/10.1063/1.328693
Turner PJ (2005) XMGRACE, Version 5.1.19. Center for coastal and land-margin research. Oregon Graduate Institute of Science and Technology, Beaverton
Amadei A, Linssen ABM, Berendsen HJC (1993) Essential dynamics of proteins. Proteins Struc Funct Genet 17(4):412–425. https://doi.org/10.1002/prot.340170408
Brown SP, Muchmore SW (2009) Large-scale application of high-throughput molecular mechanics with Poisson−Boltzmann surface area for routine physics-based scoring of protein−ligand complexes. J Med Chem 52(10):3159–3165. https://doi.org/10.1021/jm801444x
Massova I, Kollman PA (1999) Computational alanine scanning to probe protein-protein interactions: a novel approach to evaluate binding free energies. J Am Chem Soc 121(36):8133–8143. https://doi.org/10.1021/acs.jctc.7b01295
Kumari R, Kumar R, Lynn A (2014) g_mmpbsa: a GROMACS tool for high- throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex ras-raf. J Comput Chem 25(2):238–250. https://doi.org/10.1002/jcc.10379
Biswal J, Jayaprakash P, Rayala SK, Venkatraman G, Rangaswamy R, Jeyaraman J (2021) WaterMap and Molecular dynamic simulation-guided discovery of potential PAK1 inhibitors using repurposing approaches. ACS Omega 6(41):26829–26845. https://doi.org/10.1021/acsomega.1c02032
Elizabeth Sobhia M, Siva Kumar G, Sivangula S, Ghosh K, Singh H, Haokip T, Gibson J (2021) Rapid structure-based identification of potential SARS-CoV-2 main protease inhibitors. Future Med Chem 13(17):1435–1450. https://doi.org/10.4155/fmc-2020-0264
Acknowledgements
JJ thanks the TANSCHE [RGP/2019-20/ALU/HECP-0049], Indo-Taiwan [GITA/DST/TWIN/P-86/2019] dated: 04/03/2020, DBT(BIC)-No. BT/PR40154/BTIS/137/34/2021, DST-Fund for Improvement of S&T Infrastructure in Universities & Higher Educational Institutions (FIST) (SR/FST/LSI-667/2016) (C), DST Promotion of University Research and Scientific Excellence (PURSE) (No. SR/PURSE Phase 2/38 (G), 2017 and JP thanks ICMR-SRF (No: ISRM/11 (62)/2019 dated: 10/06/ 2019) to carry out this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jayaprakash, P., Biswal, J., Rangaswamy, R. et al. Designing of potent anti-diabetic molecules by targeting SIK2 using computational approaches. Mol Divers 27, 1101–1121 (2023). https://doi.org/10.1007/s11030-022-10470-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10470-0