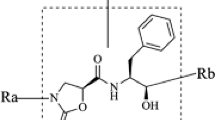
Abstract
A series of mIDH1 inhibitors derived from 3-pyrimidine-4-oxazolidin-2-ketone derivatives were studied by QSAR model to explore the key factors that inhibit mIDH1 activity. The generated model was cross-verified and non-cross-verified by Topomer CoMFA and HQSAR methods; the independent test set was verified by PLS method; the Topomer search technology was used for virtual screening and molecular design; and the Surflex-Dock method and ADMET technology were used for molecular docking, pharmacology and toxicity prediction of the designed drug molecules. The Topomer CoMFA and HQSAR cross-validation coefficients q2 are 0.783 and 0.784, respectively, and the non-cross-validation coefficients r2 are 0.978 and 0.934, respectively. Ten new drug molecules have been designed using Topomer search technology. The results of molecular docking and ADMET show that the newly designed drug molecules are effective. The docking situation, pharmacology and toxicity prediction results are good. The model can be used to predict the bioactivity of the same type of new compounds and their derivatives. The prediction results of molecular design, molecular docking and ADMET can provide some ideas for the design and development of novel mIDH1 inhibitor anticancer drugs, and provide certain theoretical basis of the experimental verification of new compounds in the future.
Graphic abstract
Newly designed molecules after docking with corresponding proteins in the PDB library, it can explore the targets of drug molecules acting with large proteins and the related force, which is very helpful for the design of new drugs and the mechanism of drug action.








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in the article and supplementary materials.
References
Popovici-Muller J, Lemieux RM, Artin E et al (2018) Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett 9(4):300–305. https://doi.org/10.1021/acsmedchemlett.7b00421
Kessler J, Güttler A, Wichmann H et al (2015) IDH1R132H mutation causes a less aggressive phenotype and radiosensitizes human malignant glioma cells independent of the oxygenation status. Radiotherapy Oncol 116(3):381–387. https://doi.org/10.1016/j.radonc.2015.08.007
Molenaar RJ, Verbaan D, Lamba S et al (2014) The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro-Oncology 16(9):1263–1273. https://doi.org/10.1093/neuonc/nou005
Li J, Huang J, Fang H et al (2016) Decreased expression of IDH1-R132H correlates with poor survival in gastrointestinal cancer. Oncotarget 7(45):73638–73650
Xuan C, Li Q, Cong W et al (2016) Prognostic and diagnostic potential of isocitrate dehydrogenase 1 in esophageal squamous cell carcinoma. Oncotarget 7(52):86148–86160
Sun N, Chen Z, Tan F et al (2013) Isocitrate dehydrogenase 1 is a novel plasma biomarker for the diagnosis of non-small cell lung cancer. Clin Cancer Res 19(18):5136–5145. https://doi.org/10.1158/1078-0432.CCR-13-0046
Gross S, Cairns RA, Minden MD et al (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Experiment Med 207(2):339–344. https://doi.org/10.1084/jem.20092506
Dang L, White DW, Gross S et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462(7274):739–744. https://doi.org/10.1038/nature08617
Xu W, Yang H, Liu Y et al (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19(1):17–30. https://doi.org/10.1016/j.ccr.2010.12.014
Parsons DW, Jones S, Zhang X et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–1812. https://doi.org/10.1126/science.1164382
Levell JR, Caferro T, Chenail G et al (2017) Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as allosteric and mutant specific inhibitors of IDH1. ACS Med Chem Lett 8(2):151–156. https://doi.org/10.1021/acsmedchemlett.6b00334
Cho YS, Levell JR, Liu G et al (2017) Discovery and evaluation of clinical candidate IDH305, a brain penetrant mutant IDH1 inhibitor. ACS Med Chem Lett 8(10):1116–1121. https://doi.org/10.1021/acsmedchemlett.7b00342
Tong J-B, Luo D, Feng Y et al (2021) Structural modification of 4, 5-dihydro-[1, 2, 4] triazolo [4, 3-f] pteridine derivatives as BRD4 inhibitors using 2D/3D-QSAR and molecular docking analysis. Molecul Divers. https://doi.org/10.1007/s11030-020-10172-5
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228. https://doi.org/10.1016/0040-4020(80)80168-2
Yao K, Liu P, Liu H et al (2019) 3D-QSAR, molecular docking and molecular dynamics simulations study of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives to explore the structure requirements of mutant IDH1 inhibitors. J Mol Struct 1189(5):187–202. https://doi.org/10.1016/j.molstruc.2019.03.092
Verma J, Khedkar V et al (2010) 3D-QSAR in drug design - a review. CTMC 10(1):95–115. https://doi.org/10.2174/156802610790232260
JitenderMarshall RG et al (1988) Three-dimensional structure-activity relationships. Trends Pharmacol Sci 9(8):285–289. https://doi.org/10.1016/0165-6147(88)90012-0
Panwar U, Singh SK (2021) Atom-based 3D-QSAR, molecular docking, DFT, and simulation studies of acylhydrazone, hydrazine, and diazene derivatives as IN-LEDGF/p75 inhibitors. Struct Chem 32(1):337–352. https://doi.org/10.1007/s11224-020-01628-3
Tenenhaus M, Vinzi VE, Chatelin YM et al (2005) PLS path modeling. Comput Stat Data Anal 48(1):159–205. https://doi.org/10.1007/s11224-020-01628-3
Shirota Y, Luo H, Qin W et al (2002) Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA Polymerase (RdRP) NS5B and modulates RNA-dependent RNA Polymerase Activity*. J Biol Chem 277(13):11149–11155. https://doi.org/10.1074/jbc.M111392200
Cramer RD, Cruz P, Stahl G et al (2009) ChemInform abstract: virtual screening for R-groups, including predicted pIC50 contributions, within large structural databases. Using Topomer CoMFA ChemInform 40(15):2180–2195. https://doi.org/10.1021/ci8001556
Ferreira MMC (2002) Multivariate QSAR. J Brazilian Chem Soc 13(6):742–753. https://doi.org/10.1590/S0103-50532002000600004
Cramer RD (2003) Topomer CoMFA: a design methodology for rapid lead optimization. J Med Chem 46(3):374–388. https://doi.org/10.1021/jm020194o
Jiao L, Zhang X, Qin Y et al (2016) Hologram QSAR study on the electrophoretic mobility of aromatic acids. Chemometrics Intell Lab Syst 157(15):202–207. https://doi.org/10.1016/j.chemolab.2016.06.020
Cheng Y, Mei Z, Tung CH et al (2010) Studies on two types of PTP1B inhibitors for the treatment of type 2 diabetes: hologram QSAR for OBA and BBB analogues. Bioorg Med Chem Lett 20(11):3329–3337. https://doi.org/10.1016/j.bmcl.2010.04.033
Waller CL (2004) A comparative QSAR study using CoMFA, HQSAR, and FRED/SKEYS paradigms for estrogen receptor binding affinities of structurally diverse compounds. J Chem Inform Comput Sci 44(2):758–765. https://doi.org/10.1021/ci0342526
Pérez-Areales FJ, Betari N, Viayna A et al (2017) Design, synthesis and multitarget biological profiling of second-generation anti-Alzheimer rhein–huprine hybrids. Future Med Chem 9(10):965–981. https://doi.org/10.4155/fmc-2017-0049
Abdizadeh R, Hadizadeh F, Abdizadeh T (2020) QSAR analysis of coumarin-based benzamides as histone deacetylase inhibitors using CoMFA, CoMSIA and HQSAR methods. J Mol Struct 1199(5):126961. https://doi.org/10.1016/j.molstruc.2019.126961
Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in drug design–a review. Curr Top Med Chem 10(1):95–115. https://doi.org/10.2174/156802610790232260
Busch KW, Swamidoss IM, Fakayode SO et al (2004) Determination of the enantiomeric composition of some molecules of pharmaceutical interest by chemometric analysis of the UV spectra of cyclodextrin guest–host complexes. Analytica Chimica Acta 525(1):53–62. https://doi.org/10.1016/j.aca.2004.07.066
Ojha PK, Mitra I, Das RN et al (2011) Further exploring rm2 metrics for validation of QSPR models. Chemometrics Intell Lab Syst 107(1):194–205. https://doi.org/10.1016/j.chemolab.2011.03.011
Rácz A, Bajusz D, Héberger K (2015) Consistency of QSAR models: correct split of training and test sets, ranking of models and performance parameters. SAR QSAR Environ Res 26(7):1–18. https://doi.org/10.1080/1062936X.2015.1084647
Golbraikh A, Tropsha A (2002) Beware of q2! J Mole Graphics Model 20(4):269–276. https://doi.org/10.1016/S1093-3263(01)00123-1
Walters WP, Stahl MT, Murcko MA (1998) Virtual screening—an overview. Drug Discovery Today 3(4):160–178. https://doi.org/10.1016/S1359-6446(97)01163-X
Doman TN, McGovern SL, Witherbee BJ et al (2002) Molecular docking and high-throughput screening for novel inhibitors of protein Tyrosine Phosphatase-1B. J Med Chem 45(11):2213–2221. https://doi.org/10.1021/jm010548w
Yuhong X, Jia S, Zhuoyong Z (2014) Topomer CoMFA and virtual screening studies of azaindole class renin inhibitors. Combinatorial Chem High Throughput Screen 17(5):458–472. https://doi.org/10.2174/1386207317666140107094708
Sterling T, Irwin JJ (2015) ZINC 15–Ligand discovery for everyone. J Chem Inf Model 55(11):2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Tong JB, Bai M, Zhao X (2016) 3D-QSAR and docking studies of HIV-1 protease inhibitors using R-group search and Surflex-dock. Med Chem Res 25(11):2619–2630. https://doi.org/10.1007/s00044-016-1701-0
Liu J, Yan L, Zhang HX et al (2012) Studies of H4R antagonists using 3D-QSAR, molecular docking and molecular dynamics. J Mol Model 18(3):991–1001. https://doi.org/10.1007/s00894-011-1137-x
Pearlman DA, Case DA, Caldwell JW et al (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1):1–41. https://doi.org/10.1016/0010-4655(95)00041-D
Cleves AE, Jain AN (2015) Knowledge-guided docking: accurate prospective prediction of bound configurations of novel ligands using Surflex-Dock. J Comput Aided Mol Des 29(6):485–509. https://doi.org/10.1007/s10822-015-9846-3
Sussman JL, Lin D, Jiang J et al (1998) Protein data bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallographica Sect D 54(61):1078–1084. https://doi.org/10.1107/S0907444998009378
Jain AN (2007) Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des 21(5):281–306
Xu C, Ren Y (2015) Molecular modeling studies of [6,6,5] Tricyclic Fused Oxazolidinones as FXa inhibitors using 3D-QSAR, Topomer CoMFA, molecular docking and molecular dynamics simulations. Bioorg Med Chem Lett 25(20):4522–4528. https://doi.org/10.1016/j.bmcl.2015.08.070
Khanna I (2012) Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discovery Today 17(19–20):1088–1102. https://doi.org/10.1016/j.drudis.2012.05.007
Cheng F, Li W, Liu G et al (2013) In silico ADMET prediction: recent advances, current challenges and future trends. Current Topics Med Chem 13(11):1273–1289. https://doi.org/10.2174/15680266113139990033
Hodgson J (2001) ADMET—turning chemicals into drugs. Nat Biotechnol 19(8):722–726. https://doi.org/10.1038/90761
Dong J, Wang N-N, Yao Z-J, et al. (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database 10(1):29. http://europepmc.org/abstract/MED/29943074https://doi.org/https://doi.org/10.1186/s13321-018-0283-xhttps://europepmc.org/articles/PMC6020094https://europepmc.org/articles/PMC6020094?pdf=render.
Artursson P, Karlsson J (1991) Correlation between oral drug absorption in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem Biophys Res Commun 175(3):880–885. https://doi.org/10.1016/0006-291X(91)91647-U
Tian S, Li Y, Wang J et al (2011) ADME evaluation in drug discovery prediction of oral bioavailability in humans based on molecular properties and structural fingerprints. J Mol Pharm 8(3):841–851
Trainor GL (2007) The importance of plasma protein binding in drug discovery. Exp Opinion Drug Discovery 2(1):51–64. https://doi.org/10.1517/17460441.2.1.51
Clark DE (2003) In silico prediction of blood–brain barrier permeation. Drug Discovery Today 8(20):927–933. https://doi.org/10.1016/S1359-6446(03)02827-7
Li J, Schneebeli ST, Bylund J et al (2011) IDSite: an accurate approach to predict P450-mediated drug metabolism. J Chem Theory Comput 7(11):3829–3845. https://doi.org/10.1021/ct200462q
Sheridan R, Korzekwa K, Torres R et al (2007) Empirical regioselectivity models for human cytochromes P450 3A4, 2D6, and 2C9. J Med Chem 50(14):3173–3184. https://doi.org/10.1021/jm0613471
Wang Y, Xing J, Xu Y et al (2015) In silico ADME/T modelling for rational drug design. Quart Rev Biophys 48(4):488–515. https://doi.org/10.1017/S0033583515000190
Aronov AM (2005) Predictive in silico modeling for hERG channel blockers. Drug Discovery Today 10(2):149–155. https://doi.org/10.1016/S1359-6446(04)03278-7
Gerald L, Kennedy et al (1986) Estimation of acute oral toxicity in rates by determination of the approximate lethal dose rather than the LD50. J Appl Toxicol 6(3):145–148. https://doi.org/10.1002/jat.2550060302
Roy PP, Leonard JT, Roy K (2008) Exploring the impact of the size of training sets for the development of predictive QSAR models. Chemom Intell Lab Sys Chemometrics Intell Lab Syst 90(1):31–42
Funding
This project was funded by the National Natural Science Foundation of China (21475081), the Natural Science Foundation of Shaanxi Province (2019JM -237), and the Graduate Innovation Fund of Shaanxi University of Science and Technology.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by Jian-Bo Tong, Shuai Bian, Xing Zhang, and Ding Luo. Data analysis was performed by Jian-Bo Tong. The first draft of the manuscript was written by Shuai Bian and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tong, JB., Bian, S., Zhang, X. et al. QSAR analysis of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives isocitrate dehydrogenase inhibitors using Topomer CoMFA and HQSAR methods. Mol Divers 26, 1017–1037 (2022). https://doi.org/10.1007/s11030-021-10222-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10222-6