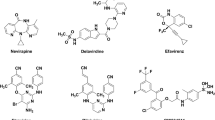
Abstract
Diarylpyrimidines (DAPYs), a type of effective HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs), have been considered as one of the most successful agents for treating AIDS. A number of structurally diverse DAPYs have been designed and synthesized in the past decade, and most of them exhibited potent anti-HIV-1 activities; however, the structure–activity relationships of recently reported DAPYs and their pharmacophore features that interacted with HIV-1 reverse transcriptase (RT) remain to be studied. In the present study, molecular docking studies were first performed on three novel classes of DAPYs to study their binding pattern in the HIV-1 RT. Based on the docking conformations of these DAPYs, 3D-QSAR models were constructed using CoMSIA and Topomer CoMFA methods, and pharmacophore models were also built using distance comparison technique. All selected DAPYs presented preferred U- or L-shaped conformations while being docked into the non-nucleoside inhibitor-binding pocket of the HIV-1 RT. The best CoMSIA model exhibited powerful predictivity, with satisfactory statistical parameters such as a q2 of 0.572, an r2 of 0.952, and an \( r_{\text{pre}}^{2} \) of 0.728. Contour maps of the best CoMSIA model were in accordance with those of the Topomer CoMFA model, giving the insight into the feature requirements of DAPYs for the anti-HIV-1 activity. Three potential pharmacophore models were constructed, and each of them was consisted of five hypothesis features. All results suggested that the aromatic ring on the left wing of DAPYs and the central pyrimidine ring contained key pharmacophore features for the anti-HIV-1 activity, and also indicated that the right wing of DAPYs had potential for further structural modification to improve activity. Eight novel DAPY molecules with potential anti-HIV-1 activities were designed on the basis of the obtained results. The findings in this study might provide important information for further design and development of novel HIV-1 NNRTIs.
Graphical Abstract










Similar content being viewed by others
References
Li W, Li X, De Clercq E, Zhan P, Liu X (2015) Discovery of potent HIV-1 non-nucleoside reverse transcriptase inhibitors from arylthioacetanilide structural motif. Eur J Med Chem 102:167–179. https://doi.org/10.1016/j.ejmech.2015.07.043
Chong P, Sebahar P, Youngman M, Garrido D, Zhang H, Stewart EL, Nolte RT, Wang L, Ferris RG, Edelstein M (2012) Rational design of potent non-nucleoside inhibitors of HIV-1 reverse transcriptase. J Med Chem 55:10601–10609. https://doi.org/10.1021/jm301294g
Esposito F, Corona A, Tramontano E (2012) HIV-1 reverse transcriptase still remains a new drug target: structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions. Mol Biol Int. https://doi.org/10.1155/2012/586401
Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA (1992) Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783–1790. https://doi.org/10.1126/science.1377403
Hsiou Y, Ding J, Das K, Clark JA, Hughes S, Arnold E (1996) Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4:853–860. https://doi.org/10.1016/S0969-2126(96)00091-3
Gu SX, Xue P, Ju XL, Zhu YY (2016) Advances in rationally designed dual inhibitors of HIV-1 reverse transcriptase and integrase. Bioorg Med Chem 24:5007–5016. https://doi.org/10.1016/j.bmc.2016.09.025
Sluiscremer N (2014) The emerging profile of cross-resistance among the nonnucleoside HIV-1 reverse transcriptase inhibitors. Viruses 6:2960–2973. https://doi.org/10.3390/v6082960
Chen X, Zhan P, Li D, De Clercq E, Liu X (2011) Recent advances in DAPYs and related analogues as HIV-1 NNRTIs. Curr Med Chem 18:359–376. https://doi.org/10.2174/092986711794839142
Sharma M, Saravolatz LD (2013) Rilpivirine: a new non-nucleoside reverse transcriptase inhibitor. J Antimicrob Chemother 68:250–256. https://doi.org/10.1093/jac/dks404
Das K, Bauman JD, Clark AD Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E (2008) High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA 105:1466–1471. https://doi.org/10.1073/pnas.0711209105
Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E (2009) Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol 385:693–713. https://doi.org/10.1016/j.jmb.2008.10.071
Liang YH, He QQ, Zeng ZS, Liu ZQ, Feng XQ, Chen FE, Balzarini J, Pannecouque C, De Clercq E (2010) Synthesis and anti-HIV activity of 2-naphthyl substituted DAPY analogues as non-nucleoside reverse transcriptase inhibitors. Bioorg Med Chem 18:4601–4605. https://doi.org/10.1016/j.bmc.2010.05.036
Wan ZY, Yao J, Mao TQ, Wang XL, Wang HF, Chen WX, Yin H, Chen FE, De Clercq E, Daelemans D, Pannecouque C (2015) Pyrimidine sulfonylacetanilides with improved potency against key mutant viruses of HIV-1 by specific targeting of a highly conserved residue. Eur J Med Chem 102:215–222. https://doi.org/10.1016/j.ejmech.2015.08.007
Kertesz DJ, Brotherton-Pleiss C, Yang M, Wang Z, Lin X, Qiu Z, Hirschfeld DR, Gleason S, Mirzadegan T, Dunten PW (2010) Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. -Benzyl derivatives with broad potency against resistant mutant viruses. Bioorg Med Chem Lett 20:6020–6023. https://doi.org/10.1016/j.bmcl.2010.05.040
Liu X, Chen XW, Zhang LZ, Zhan P, Liu XY (2015) 3D-QSAR and docking studies on piperidine-substituted diarylpyrimidine analogues as HIV-1 reverse transcriptase inhibitors. Med Chem Res 24:3314–3326. https://doi.org/10.1007/s00044-015-1381-1
Wu HQ, Yao J, He QQ, Chen FE (2014) Docking-based CoMFA and CoMSIA studies on naphthyl-substituted diarylpyrimidines as NNRTIs. SAR QSAR Environ Res 25:761–775. https://doi.org/10.1080/1062936X.2014.955054
Peddi SR, Mohammed NA, Hussein AA, Sivan SK (2018) Multiple-receptor conformation docking, dock pose clustering, and 3D QSAR-driven approaches exploring new HIV-1 RT inhibitors. Struct Chem. https://doi.org/10.1007/s11224-018-1082-8
Ma XD, Yang SQ, Gu SX, He QQ, Chen FE, De Clercq E, Balzarini J, Pannecouque C (2011) Synthesis and anti-HIV activity of aryl-2-[(4-cyanophenyl)amino]-4-pyrimidinone hydrazones as potent non-nucleoside reverse transcriptase inhibitors. ChemMedChem 6:2225–2232. https://doi.org/10.1002/cmdc.201100334
Wan ZY, Yao J, Tao Y, Mao TQ, Wang XL, Lu YP, Wang HF, Yin H, Wu Y, Chen FE, De Clercq E, Daelemans D, Pannecouque C (2015) Discovery of piperidin-4-yl-aminopyrimidine derivatives as potent non-nucleoside HIV-1 reverse transcriptase inhibitors. Eur J Med Chem 97:1–9. https://doi.org/10.1016/j.ejmech.2015.04.050
Mouchlis VD, Mavromoustakos TM, Kokotos G (2010) Design of new secreted phospholipase A2 inhibitors based on docking calculations by modifying the pharmacophore segments of the FPL67047XX inhibitor. J Comput Aided Mol Des 24:107–115. https://doi.org/10.1007/s10822-010-9319-7
Ruppert J, Welch W, Jain AN (1997) Automatic identification and representation of protein binding sites for molecular docking. Protein Sci 6:524–533. https://doi.org/10.1002/pro.5560060302
Perola E, Charifson PS (2004) Conformational analysis of drug-like molecules bound to proteins: an extensive study of ligand reorganization upon binding. J Med Chem 47:2499–2510. https://doi.org/10.1021/jm030563w
Nicklaus MC, Wang SM, Driscoll JS, Milne GWA (1995) Conformational changes of small molecules binding to proteins. Bioorg Med Chem 3:411–428. https://doi.org/10.1016/0968-0896(95)00031-B
Foloppe N, Chen IJ (2009) Conformational sampling and energetics of drug-like molecules. Curr Med Chem 16:3381–3413. https://doi.org/10.2174/092986709789057680
Avgy-david HH, Senderowitz H (2015) Toward focusing conformational ensembles on bioactive conformations: a molecular mechanics/quantum mechanics study. J Chem Inf Model 55(10):2154–2167. https://doi.org/10.1021/acs.jcim.5b00259
Liu GY, Ju XL, Cheng J (2010) Selectivity of Imidacloprid for fruit fly versus rat nicotinic acetylcholine receptors by molecular modeling. J Mol Model 16:993–1002. https://doi.org/10.1007/s00894-009-0601-3
Cramer RD III, Bunce JD, Patterson DE, Frank IE (1988) Crossvalidation, bootstrapping, and partial least squares compared with multiple regression in conventional QSAR studies. Mol Inform 7:18–25. https://doi.org/10.1002/qsar.19880070105
Romero-Parra J, Chung H, Tapia RA, Faúndez M, Morales-Verdejo C, Lorca M, Lagos CF, Di Marzo V, David Pessoa-Mahana C, Mella J (2017) Combined CoMFA and CoMSIA 3D-QSAR study of benzimidazole and benzothiophene derivatives with selective affinity for the CB2 cannabinoid receptor. Eur J Pharm Sci 101:1–10. https://doi.org/10.1016/j.ejps.2017.01.037
Golbraikh A, Tropsha A (2002) Beware of q 2! J Mol Graph Mod 20:269–276. https://doi.org/10.1016/S1093-3263(01)00123-1
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring r 2m metrics for validation of QSPR models. Chemom Intell Lab 107:194–205. https://doi.org/10.1016/j.chemolab.2011.03.011
Alexander DLJ, Tropsha A, Winkler DA (2015) Beware of R2: simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J Chem Inf Model 55:1316–1322. https://doi.org/10.1021/acs.jcim.5b00206
Cramer RD (2003) Topomer CoMFA: a design methodology for rapid lead optimization. J Med Chem 46:374–388. https://doi.org/10.1021/jm020194o
Cramer RD, Clark RD, Patterson DE, Ferguson AM (1996) Bioisosterism as a molecular diversity descriptor: steric fields of single “topomeric” conformers. J Med Chem 39:3060–3069. https://doi.org/10.1021/jm960291f
Tucker TJ, Saggar S, Sisko JT, Tynebor RM, Williams TM, Felock PJ, Flynn JA, Lai MT, Liang Y, McGaughey G, Liu M, Miller M, Moyer G, Munshi V, Perlow-Poehnelt R, Prasad S, Sanchez R, Torrent M, Vacca JP, Wan BL, Yan Y (2008) The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations. Bioorg Med Chem Lett 18:2959–2966. https://doi.org/10.1016/j.bmcl.2008.03.064
Yusuf D, Davis AM, Kleywegt GJ, Schmitt S (2008) An alternative method for the evaluation of docking performance: RSR versus RMSD. J Chem Inf Model 48:1411–1422. https://doi.org/10.1021/ci800084x
Das K, Arnold E (2013) HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr Opin Virol 3:119–128. https://doi.org/10.1016/j.coviro.2013.03.012
Böhm M, Strzebecher J, Klebe G (1999) Three-dimensional quantitative structure-activity relationship analyses using comparative molecular field analysis and comparative molecular similarity indices analysis to elucidate selectivity differences of inhibitors binding to trypsin, thrombin, and factor Xa. J Med Chem 42:458–477. https://doi.org/10.1021/jm981062r
Roy K, Kar S, Das RN (2015) Statistical methods in QSAR/QSPR. In: A primer on QSAR/QSPR modeling. SpringerBriefs in Molecular Science. Springer, Cham, pp 37-59. https://doi.org/10.1007/978-3-319-17281-1
Liu GY, Ju XL, Cheng J, Liu ZQ (2010) 3D-QSAR studies of insecticidal anthranilic diamides as ryanodine receptor activators using CoMFA, CoMSIA and DISCOtech. Chemosphere 78:300–306. https://doi.org/10.1016/j.chemosphere.2009.10.038
Acknowledgements
This work was supported by Graduate Innovative Fund of Wuhan Institute of Technology (No. CX2017113), Natural Science Foundation of Hubei Province of China (No. 2017CFB121), Hubei Provincial Department of Education of China (No. Q20171503), Wuhan International Scientific and Technological Cooperation Project (No. 2017030209020257), and National Natural Science Foundation of China (No. 21402148).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, G., Wan, Y., Wang, W. et al. Docking-based 3D-QSAR and pharmacophore studies on diarylpyrimidines as non-nucleoside inhibitors of HIV-1 reverse transcriptase. Mol Divers 23, 107–121 (2019). https://doi.org/10.1007/s11030-018-9860-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-018-9860-1