Abstract
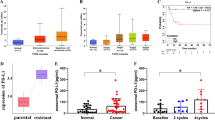
Previous studies have reported that exosomes bearing certain microRNAs (miRNAs) are related to the physiological functions of different types of cancer cells. Our study aimed to elucidate the role of miR-200a in esophageal squamous cell carcinoma (ESCC). We observed that miR-200a expression is higher in esophageal carcinoma cells, tissues, and exosomes than in normal cells and healthy tissues. We showed that exosome-shuttled miR-200a promotes the proliferation, migration, and invasion of esophageal cells and inhibits apoptosis, thereby leading to the progression of ESCC. We showed that miR-200a exerts its effects through its interaction with Keap1, thus altering the Keap1/Nrf2 signaling pathway. Our results suggest that exosome-shuttled miR-200a might be useful as a biomarker for prognosis in patients with ESCC.






Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Enzinger PC, Mayer RJ (2003) Esophageal cancer. N Engl J Med 349(23):2241–2252
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin D, Piñeros M et al (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144(8):1941–1953
Kollarova H, Machova L, Horakova D, Janoutova G, Janout V (2007) Epidemiology of esophageal cancer-an overview article. Biomed Pap Med Fac Palacky Univ Olomouc. https://doi.org/10.5507/bp.2007.003
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101(10):2087–2092
Mäkitie RE, Hackl M, Niinimäki R, Kakko S, Grillari J, Mäkitie O (2018) Altered MicroRNA profile in osteoporosis caused by impaired WNT signaling. J Clin Endocrinol Metab 103(5):1985–1996
Chen L, Sun DZ, Fu YG, Yang PZ, Lv HQ, Gao Y, Zhang XY (2020) Upregulation of microRNA-141 suppresses epithelialmesenchymal transition and lymph node metastasis in laryngeal cancer through HOXC6-dependent TGF-β signaling pathway. Cell Signal 66:109444. https://doi.org/10.1016/j.cellsig.2019.109444
Yang J, Hu F, Fu X, Jiang Z, Zhang W, Chen K (2019) MiR-128/SOX7 alleviates myocardial ischemia injury by regulating IL-33/sST2 in acute myocardial infarction. Biol Chem 400(4):533–544
Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodriguez-Dorantes M (2014) MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther 7:1327
Ramachandran S, Palanisamy V (2012) Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip Rev 3(2):286–293
Kosaka N, Yusuke Y, Hagiwara K, Tominaga N, Katsuda T, Ochiya T (2013) Trash or treasure: extracellular microRNAs and cell-to-cell communication. Front Genet 4:173
Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10(1):42–46
Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110(1):13–21
Saydam O, Shen Y, Würdinger T, Senol O, Boke E, James MF et al (2009) Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway. Mol Cell Biol 29(21):5923–5940
Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T et al (2012) MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin. Int J Oncol 40(4):1162–1170
Yu S-J, Hu J-Y, Kuang X-Y, Luo J-M, Hou Y-F, Di G-H et al (2013) MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res 19(6):1389–1399
Yuan Jh, Yang F, Chen Bf LuZ, Xs Huo, Wp Zhou et al (2011) The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 54(6):2025–2035
Cong N, Du P, Zhang A, Shen F, Su J, Pu P et al (2013) Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep 29(4):1579–1587
Wu Q, Guo R, Lin M, Zhou B, Wang Y (2011) MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol 122(1):149–154
Eades G, Yang M, Yao Y, Zhang Y, Zhou Q (2011) miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286(47):40725–40733
Han Y, Wei F, Xu X, Cai Y, Chen B, Wang J, et al. (2002) Establishment and comparative genomic hybridization analysis of human esophageal carcinomas cell line EC9706. Zhonghua yi xue yi chuan xue za zhi= Zhonghua yixue yichuanxue zazhi= Chinese journal of medical genetics 19(6):455.
Yang M, Chen J, Su F, Yu B, Su F, Lin L et al (2011) Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 10(1):117
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife. https://doi.org/10.7554/eLife.05005
Friedman RC, Farh KK-H, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Wu F, Li M, You W, Ji Y, Cui X, Hu J et al (2018) A genetic variant in miR-124 decreased the susceptibility to esophageal squamous cell carcinoma in a Chinese Kazakh population. Genet Test Mol Biomark 22(1):29–34
Zhang J, Jiao Q, Kong L, Yu J, Fang A, Li M et al (2018) Nrf2 and Keap1 abnormalities in esophageal squamous cell carcinoma and association with the effect of chemoradiotherapy. Thoracic cancer 9(6):726–735
Sato Y, Kobayashi H, Suto Y, Olney H, Davis E, Super H et al (2001) Chromosomal instability in chromosome band 12p13: multiple breaks leading to complex rearrangements including cytogenetically undetectable sub-clones. Leukemia 15(8):1193–1202
Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q (2011) miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem 286(29):25992–26002
Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF et al (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Can Res 68(19):7846–7854
Uhlmann S, Zhang J, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S et al (2010) miR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29(30):4297–4306
Zhen Q, Liu J, Gao L, Liu J, Wang R, Chu W et al (2015) MicroRNA-200a targets EGFR and c-Met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. Cytogenet Genome Res 146(1):1–8
Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L et al (2015) miR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep 33(2):713–720
Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian X-W et al (2010) miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun 391(1):535–541
Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P et al (2015) Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol 17(10):779–787
Jia C, Zhang Y, Xie Y, Ren Y, Zhang H, Zhou Y et al (2019) miR-200a-3p plays tumor suppressor roles in gastric cancer cells by targeting KLF12. Artif Cells, Nanomed Biotechnol 47(1):3697–3703
Borek C (2004) Antioxidants and radiation therapy. J Nutr 134(11):3207S-S3209
Ozben T (2007) Oxidative stress and apoptosis: impact on cancer therapy. J Pharm Sci 96(9):2181–2196
Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T et al (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24(16):7130–7139
Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M (2001) Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6(10):857–868
Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y et al (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem 275(21):16023–16029
Stacy DR, Ely K, Massion PP, Yarbrough WG, Hallahan DE, Sekhar KR et al (2006) Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head & Neck 28(9):813–818
Lee S, Lim M-J, Kim M-H, Yu C-H, Yun Y-S, Ahn J et al (2012) An effective strategy for increasing the radiosensitivity of human lung cancer cells by blocking Nrf2-dependent antioxidant responses. Free Radical Biol Med 53(4):807–816
Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ et al (2017) Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov 7(1):86–101
Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K et al (2011) NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 13(9):864-IN26
Eades G, Yang M, Yao Y, Zhang Y, Zhou Q (2011) miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells*. J Biol Chem 286(47):40725–40733
Jiang Z, Wu J, Ma F, Jiang J, Xu L, Du L et al (2020) MicroRNA-200a improves diabetic endothelial dysfunction by targeting KEAP1/NRF2. J Endocrinol 245(1):129–140
Wang X, Ye L, Zhang K, Gao L, Xiao J, Zhang Y (2020) Upregulation of microRNA-200a in bone marrow mesenchymal stem cells enhances the repair of spinal cord injury in rats by reducing oxidative stress and regulating Keap1/Nrf2 pathway. Artif Organs 44(7):744–752
Ma Y, Pan C, Tang X, Zhang M, Shi H, Wang T, et al. (2020) MicroRNA-200a represses myocardial infarction-related cell death and inflammation by targeting the Keap1/Nrf2 and β-catenin pathways. Hellenic journal of cardiology. HJC = Hellenike kardiologike epitheorese.
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
PL, XL, and WX conceived and designed the research. PL, XL, WX, HQ, RL, SL, and HS performed experiments, interpreted the results of experiments, and analyzed data. PL edited and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital and written informed consent was obtained from all patients. All animal experiments were performed in accordance with the International Commission Guidelines for the Care and Use of Laboratory Animals.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2022_4353_MOESM1_ESM.tif
Supplementary file1 (TIF 7422 kb) Exosome isolated from EC9706 cells and HEECs. TEM images of exosomes derived from EC9706 cells and HEECs
11010_2022_4353_MOESM2_ESM.tif
Supplementary file2 (TIF 113 kb) MiR-200a level in purified exosome from EC9706 cells and HEECs. qPCR was carried out to examine the miR-200a level in purified exosome from EC9706 cells and HEECs
11010_2022_4353_MOESM3_ESM.tif
Supplementary file3 (TIF 185 kb) Effect of co-culture system on the miR-200a level in donor EC9706 cells, purified exosomes, and recipient EC9706 cells. Donor EC9706 cells were transfected with miR-200a mimic/inhibitor (100 nM) or NC mimic/inhibitor (100 nM) for 24 h and then subjected to co-culture system. The exosomes derived from donor cells were also purified. (A, B, C) qPCR quantification of miR-200a levels in donor ESCC EC9706 cells, exosomes, and recipient EC9706 cells. * p < 0.05, ** p < 0.01, *** p < 0.001. Data are shown as mean ± standard deviation, and one-way ANOVA and Tukey’s post hoc test were used to compare multiple groups. Experiments were performed three times
11010_2022_4353_MOESM4_ESM.tif
Supplementary file4 (TIF 11726 kb) Co-culture system affected the proliferation, apoptosis, migration, and invasion of EC9706 cells and reduced apoptosis. (A) EdU staining assay to assess the proliferation of recipient ESCC EC9706 cells. (B) CFA to examine the colony formation ability of recipient cells. (C) FC assay to detect apoptosis of recipient EC9706 cells in each group. (D) Wound healing assay to assess the migration of recipient EC9706 cells in two groups. (E) Transwell assay to examine the invasion of recipient EC9706 cells in two groups. Donor EC9706 cells transfected with a miR-200a overexpression or inhibitor construct were co-cultured with recipient EC9706 cells. * p < 0.05, ** p < 0.01. Data are shown as mean ± standard deviation, and one-way ANOVA and Tukey’s post hoc test were used to compare multiple groups. Experiments were performed three times
Rights and permissions
About this article
Cite this article
Li, P., Liu, X., Xing, W. et al. Exosome-derived miR-200a promotes esophageal cancer cell proliferation and migration via the mediating Keap1 expression. Mol Cell Biochem 477, 1295–1308 (2022). https://doi.org/10.1007/s11010-022-04353-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04353-z