Abstract
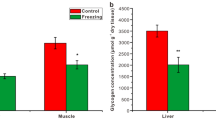
The wood frog, Rana sylvatica, is one of only a few vertebrate species that display natural freeze tolerance. Frogs survive the freezing of about two-thirds of their body water as extracellular ice over the winter months. Multiple adaptations support freeze tolerance including metabolic rate depression and the production of huge amounts of glucose (often 200 mM or more) as a cryoprotectant that protects cells from freeze damage. To understand how high glucose levels affect gene expression, we studied MondoA, a glucose sensing transcription factor, and its partner MLX (Max-like protein) to assess their ability to modulate the expression of genes involved in glucose metabolism and circadian rhythm. Wood frog liver and brain tissues were analyzed, assessing protein levels, nuclear distribution, and DNA binding activity of MondoA:MLX during freezing (24 h at − 2.5 °C) and subsequent thawing (8 h returned to 5 °C), as compared with 5 °C controls. Downstream targets of MondoA:MLX were also evaluated: TXNIP (thioredoxin interacting protein), ARRDC4 (arrestin domain containing 4), HK-2 (hexokinase-2), PFKFB-3 (6-phosphofructo-2-kinase isozyme 3) and KLF-10 (Kruppel-like factor-10). Both KLF-10 and PFKFB-3 are also involved in circadian dependant regulation which was also explored in the current study via analysis of BMAL-1 (aryl hydrocarbon receptor nuclear translocator-like protein 1) and CLOCK (circadian locomotor output cycles kaput) proteins. Our data establish the MondoA-MLX complex as active under the hyperglycemic conditions in liver to regulate glucose metabolism and may also link to circadian rhythm in liver via KLF-10 and PFKFB-3 but not in brain.









Similar content being viewed by others
References
Storey KB, Storey JM (1996) Natural freezing survival in animals. Annu Rev Ecol Syst 27:365–386. https://doi.org/10.1146/annurev.ecolsys.27.1.365
Storey KB, Storey JM (2017) Molecular physiology of freeze tolerance in vertebrates. Physiol Rev 97:623–665. https://doi.org/10.1152/physrev.00016.2016
Layne J, Lee R (1995) Adaptations of frogs to survive freezing. Clim Res 5:53–59. https://doi.org/10.3354/cr005053
Costanzo JP, Lee RE (2013) Avoidance and tolerance of freezing in ectothermic vertebrates. J Exp Biol 216:1961–1967. https://doi.org/10.1242/jeb.070268
Costanzo JP, Lee RE (1996) Mini-review: ice nucleation in freeze-tolerant vertebrates. Cryoletters 17(2):111–118
Storey KB, Storey JM (1984) Biochemical adaption for freezing tolerance in the wood frog, Rana sylvatica. J Comp Physiol B 155:29–36. https://doi.org/10.1007/BF00688788
Rosendale AJ, Lee RE, Costanzo JP (2014) Effect of physiological stress on expression of glucose transporter 2 in liver of the wood frog, Rana sylvatica. J Exp Zool A 321:566–576. https://doi.org/10.1002/jez.1885
Storey KB (2000) Vertebrate freeze tolerance: molecular studies of signal transduction and gene expression. In: Heldmaier G, Klaus S, Klingenspor M (eds) Life in the cold. Springer, Berlin, pp 527–539
Storey K, Storey J (2004) Physiology, biochemistry, and molecular biology of vertebrate freeze tolerance. In: Benson E, Fuller B, Lane N (eds) Life in the frozen state. CRC Press, Boca Raton, pp 243–274
Zhang J, Storey KB (2013) Akt signaling and freezing survival in the wood frog, Rana sylvatica. Biochim Biophys Acta Gen Subj 1830:4828–4837. https://doi.org/10.1016/j.bbagen.2013.06.020
Lambert SA, Jolma A, Campitelli LF et al (2018) The human transcription factors. Cell 172:650–665. https://doi.org/10.1016/j.cell.2018.01.029
Brand LH, Kirchler T, Hummel S et al (2010) DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6:25. https://doi.org/10.1186/1746-4811-6-25
Al-Attar R, Storey KB (2018) Effects of anoxic exposure on the nuclear factor of activated T cell (NFAT) transcription factors in the stress-tolerant wood frog. Cell Biochem Funct 36:420–430. https://doi.org/10.1002/cbf.3362
Sans CL, Satterwhite DJ, Stoltzman CA et al (2006) MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol 26:4863–4871. https://doi.org/10.1128/MCB.00657-05
Wilde BR, Ayer DE (2015) Interactions between Myc and MondoA transcription factors in metabolism and tumourigenesis. Br J Cancer 113:1529–1533. https://doi.org/10.1038/bjc.2015.360
Richards P, Rachdi L, Oshima M et al (2018) MondoA is an essential glucose-responsive transcription factor in human pancreatic β-cells. Diabetes 67:461–472. https://doi.org/10.2337/db17-0595
Stoltzman CA, Peterson CW, Breen KT et al (2008) Glucose sensing by MondoA: Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA 105:6912–6917. https://doi.org/10.1073/pnas.0712199105
Vazquez Illanes MD, Storey KB (1993) 6-Phosphofructo-2-kinase and control of cryoprotectant synthesis in freeze tolerant frogs. Biochim Biophys Acta 1158:29–32. https://doi.org/10.1016/0304-4165(93)90092-m
Mattila J, Hietakangas V (2017) Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 207:1231–1253. https://doi.org/10.1534/genetics.117.199885
Kalsbeek A, la Fleur S, Fliers E (2014) Circadian control of glucose metabolism. Mol Metab 3:372–383. https://doi.org/10.1016/j.molmet.2014.03.002
Qian J, Scheer FAJL (2016) Circadian system and glucose metabolism: implications for physiology and disease. Trends Endocrinol Metab 27:282–293. https://doi.org/10.1016/j.tem.2016.03.005
Gachon F, Loizides-Mangold U, Petrenko V, Dibner C (2017) Glucose homeostasis: regulation by peripheral circadian clocks in rodents and humans. Endocrinology 158:1074–1084. https://doi.org/10.1210/en.2017-00218
Lande-Diner L, Boyault C, Kim JY, Weitz CJ (2013) A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA 110:16021–16026. https://doi.org/10.1073/pnas.1305980110
Farhud D, Aryan Z (2018) Circadian rhythm, lifestyle and health: a narrative review. Iran J Public Health 47:1068–1076
Ahn B, Wan S, Jaiswal N et al (2019) MondoA drives muscle lipid accumulation and insulin resistance. JCI Insight 4:1–16. https://doi.org/10.1172/jci.insight.129119
Havula E, Teesalu M, Hyötyläinen T et al (2013) Mondo/ChREBP-Mlx-regulated transcriptional network is essential for dietary sugar tolerance in Drosophila. PLoS Genet 9:e1003438. https://doi.org/10.1371/journal.pgen.1003438
Guillaumond F, Grechez-Cassiau A, Subramaniam M et al (2010) Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol 30:3059–3070. https://doi.org/10.1128/MCB.01141-09
Kaczynski J, Cook T, Urrutia R (2003) Sp1- and Krüppel-like transcription factors. Genome Biol 4(2):206
Chen L, Zhao J, Tang Q et al (2016) PFKFB3 control of cancer growth by responding to circadian clock outputs. Sci Rep 6:24324. https://doi.org/10.1038/srep24324
Krivoruchko A, Storey KB (2014) Activation of the carbohydrate response element binding protein (ChREBP) in response to anoxia in the turtle Trachemys scripta elegans. Biochim Biophys Acta Gen Subj 1840:3000–3005. https://doi.org/10.1016/j.bbagen.2014.06.001
Gerber VEM, Wijenayake S, Storey KB (2016) Anti-apoptotic response during anoxia and recovery in a freeze-tolerant wood frog (Rana sylvatica). PeerJ 4:e1834. https://doi.org/10.7717/peerj.1834
Eaton SL, Roche SL, Llavero Hurtado M et al (2013) Total protein analysis as a reliable loading control for quantitative fluorescent western blotting. PLoS ONE 8:e72457. https://doi.org/10.1371/journal.pone.0072457
Zhang J, Storey KB (2016) RBioplot: an easy-to-use R pipeline for automated statistical analysis and data visualization in molecular biology and biochemistry. PeerJ 4:e2436. https://doi.org/10.7717/peerj.2436
Storey KB, Storey JM (1986) Freeze tolerant frogs: cryoprotectants and tissue metabolism during freeze–thaw cycles. Can J Zool 64:49–56. https://doi.org/10.1139/z86-008
Kaadige MR, Yang J, Wilde BR, Ayer DE (2015) MondoA-Mlx transcriptional activity is limited by mTOR-MondoA interaction. Mol Cell Biol 35:101–110. https://doi.org/10.1128/MCB.00636-14
Hadj-Moussa H, Storey KB (2018) Micromanaging freeze tolerance: the biogenesis and regulation of neuroprotective microRNAs in frozen brains. Cell Mol Life Sci 75:3635–3647. https://doi.org/10.1007/s00018-018-2821-0
O’Donnell AF, Schmidt MC (2019) AMPK-mediated regulation of alpha-arrestins and protein trafficking. Int J Mol Sci 20:515. https://doi.org/10.3390/ijms20030515
Han K-S, Ayer DE (2013) MondoA senses adenine nucleotides: transcriptional induction of thioredoxin-interacting protein. Biochem J 453:209–218. https://doi.org/10.1042/BJ20121126
Chutkow WA, Patwari P, Yoshioka J, Lee RT (2008) Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J Biol Chem 283:2397–2406. https://doi.org/10.1074/jbc.M708169200
Alhawiti NM, Al Mahri S, Aziz MA et al (2017) TXNIP in metabolic regulation: physiological role and therapeutic outlook. Curr Drug Targets 18:1095–1103. https://doi.org/10.2174/1389450118666170130145514
King PA, Rosholt MN, Storey KB (1995) Seasonal changes in plasma membrane glucose transporters enhance cryoprotectant distribution in the freeze-tolerant wood frog. Can J Zool 73:1–9. https://doi.org/10.1139/z95-001
Dotimas JR, Lee AW, Schmider AB et al (2016) Diabetes regulates fructose absorption through thioredoxin-interacting protein. eLife 5:e18313. https://doi.org/10.7554/eLife.18313
Nasoohi S, Ismael S, Ishrat T (2018) Thioredoxin-interacting protein (TXNIP) in cerebrovascular and neurodegenerative diseases: regulation and implication. Mol Neurobiol 55:7900–7920. https://doi.org/10.1007/s12035-018-0917-z
Hou Y, Wang Y, He Q et al (2018) Nrf2 inhibits NLRP3 inflammasome activation through regulating Trx1/TXNIP complex in cerebral ischemia reperfusion injury. Behav Brain Res 336:32–39. https://doi.org/10.1016/j.bbr.2017.06.027
Wu C-W, Tessier SN, Storey KB (2018) Stress-induced antioxidant defense and protein chaperone response in the freeze-tolerant wood frog Rana sylvatica. Cell Stress Chaperones 23:1205–1217. https://doi.org/10.1007/s12192-018-0926-x
Mochizuki M, Kwon Y-W, Yodoi J, Masutani H (2009) Thioredoxin regulates cell cycle via the ERK1/2-Cyclin D1 Pathway. Antioxid Redox Signal 11:2957–2971. https://doi.org/10.1089/ars.2009.2623
Zhang J, Storey KB (2012) Cell cycle regulation in the freeze tolerant wood frog, Rana sylvatica. Cell Cycle 11:1727–1742. https://doi.org/10.4161/cc.19880
Patwari P, Lee RT (2012) An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab 23:216–222. https://doi.org/10.1016/j.tem.2012.03.003
Kolar D, Gresikova M, Waskova-Arnostova P et al (2017) Adaptation to chronic continuous hypoxia potentiates Akt/HK2 anti-apoptotic pathway during brief myocardial ischemia/reperfusion insult. Mol Cell Biochem 432:99–108. https://doi.org/10.1007/s11010-017-3001-5
Joanisse DR, Storey KB (1996) Oxidative damage and antioxidants in Rana sylvatica, the freeze-tolerant wood frog. Am J Physiol Integr Comp Physiol 271:R545–R553. https://doi.org/10.1152/ajpregu.1996.271.3.R545
Peng H, Zhu Y, Goldberg J et al (2019) DNA methylation of five core circadian genes jointly contributes to glucose metabolism: a gene-set analysis in monozygotic twins. Front Genet 10:1–7. https://doi.org/10.3389/fgene.2019.00329
Aschoff J, Fatranska M, Giedke H et al (1971) Human circadian rhythms in continuous darkness: entrainment by social cues. Science (80-) 171:213–215. https://doi.org/10.1126/science.171.3967.213
Dibner C, Schibler U (2015) Circadian timing of metabolism in animal models and humans. J Intern Med 277:513–527. https://doi.org/10.1111/joim.12347
Memon A, Lee W (2018) KLF10 as a tumor suppressor gene and its TGF-β signaling. Cancers (Basel) 10:161. https://doi.org/10.3390/cancers10060161
Papadakis KA, Krempski J, Reiter J et al (2015) Krüppel-like factor KLF10 regulates transforming growth factor receptor II expression and TGF-β signaling in CD8+ T lymphocytes. Am J Physiol 308:C362–C371. https://doi.org/10.1152/ajpcell.00262.2014
Aguilar OA, Hadj-Moussa H, Storey KB (2016) Regulation of SMAD transcription factors during freezing in the freeze tolerant wood frog, Rana sylvatica. Comp Biochem Physiol B 201:64–71. https://doi.org/10.1016/j.cbpb.2016.07.003
Michael AK, Fribourgh JL, Chelliah Y et al (2017) Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci USA 114:1560–1565. https://doi.org/10.1073/pnas.1615310114
Mehra A, Baker CL, Loros JJ, Dunlap JC (2009) Post-translational modifications in circadian rhythms. Trends Biochem Sci 34:483–490. https://doi.org/10.1016/j.tibs.2009.06.006
Subramaniam M, Hawse JR, Rajamannan NM et al (2010) Functional role of KLF10 in multiple disease processes. BioFactors 36:8–18. https://doi.org/10.1002/biof.67
Storey KB, Storey JM (2019) Mitochondria, metabolic control and microRNA: advances in understanding amphibian freeze tolerance. BioFactors. https://doi.org/10.1002/biof.1511
Greenway SC, Storey KB (2000) Activation of mitogen-activated protein kinases during natural freezing and thawing in the wood frog. Mol Cell Biochem 209:29–37
Kennaway DJ, Varcoe TJ, Voultsios A, Boden MJ (2013) Global loss of bmal1 expression alters adipose tissue hormones, gene expression and glucose metabolism. PLoS ONE 8:e65255. https://doi.org/10.1371/journal.pone.0065255
Acknowledgements
Research was supported by a Discovery Grant to KBS from the Natural Sciences and Engineering Research Council of Canada (#6793); KBS holds the Canada Research Chair in Molecular Physiology. The authors thank J.M. Storey for editorial review of manuscript, and R. Al-Attar for providing technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, G., Storey, K.B. MondoA:MLX complex regulates glucose-dependent gene expression and links to circadian rhythm in liver and brain of the freeze-tolerant wood frog, Rana sylvatica. Mol Cell Biochem 473, 203–216 (2020). https://doi.org/10.1007/s11010-020-03820-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03820-9