Abstract
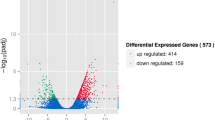
When temperatures plummet below 0 °C, wood frogs (Rana sylvatica) can endure the freezing of up to ~ 65% of their body water in extracellular ice masses, displaying no measurable brain activity, no breathing, no movement, and a flat-lined heart. To aid survival, frogs retreat into a state of suspended animation characterized by global suppression of metabolic functions and reprioritization of energy usage to essential survival processes that is elicited, in part, by the regulatory controls of microRNAs. The present study is the first to investigate miRNA biogenesis and regulation in the brain of a freeze tolerant vertebrate. Indeed, proper brain function and adaptations to environmental stimuli play a crucial role in coordinating stress responses. Immunoblotting of miRNA biogenesis factors illustrated an overall reduction in the majority of these processing proteins suggesting a potential suppression of miRNA maturation over the freeze–thaw cycle. This was coupled with a large-scale RT-qPCR analysis of relative expression levels of 113 microRNA species in the brains of control, 24 h frozen, and 8 h thawed R. sylvatica. Of the 41 microRNAs differentially regulated during freezing and thawing, only two were significantly upregulated. Bioinformatic target enrichment of the downregulated miRNAs, performed at the low temperatures experienced during freezing and thawing, predicted their involvement in the potential activation of various neuroprotective processes such as synaptic signaling, intracellular signal transduction, and anoxia/ischemia injury protection. The predominantly downregulated microRNA fingerprint identified herein suggests a microRNA-mediated cryoprotective mechanism responsible for maintaining neuronal functions and facilitating successful whole brain freezing and thawing.




Similar content being viewed by others
References
Costanzo JP, Lee RE, Ultsch GR (2008) Physiological ecology of overwintering in hatchling turtles. J Exp Zool Part A Ecol Genet Physiol 309A:297–379
Denlinger DL, Lee RE (2010) Low temperature biology of insects. Cambridge University Press, Cambridge
Holmstrup M (2014) The ins and outs of water dynamics in cold tolerant soil invertebrates. J Therm Biol 45:117–123
Strimbeck GR, Schaberg PG, Fossdal CG et al (2015) Extreme low temperature tolerance in woody plants. Front Plant Sci 6:884
Storey KB, Storey JM (2017) Molecular physiology of freeze tolerance in vertebrates. Physiol Rev 97:623–665
Storey KB (1990) Life in a frozen state: adaptive strategies for natural freeze tolerance in amphibians and reptiles. Am J Physiol 258:R559–R568
Dawson NJ, Katzenback BA, Storey KB (2015) Free-radical first responders: the characterization of CuZnSOD and MnSOD regulation during freezing of the freeze-tolerant North American wood frog, Rana sylvatica. Biochim Biophys Acta Gen Subj 1850:97–106
Storey KB, Storey JM (2004) Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev Camb Philos Soc 79:207–233
Storey KB (2006) Reptile freeze tolerance: metabolism and gene expression. Cryobiology 52:1–16
Storey KB (2015) Regulation of hypometabolism: insights into epigenetic controls. J Exp Biol 218:150–159
Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Biggar KK, Storey KB (2011) The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol 3:167–175
Leung AKL, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40:205–215
Wu P, Nakano S, Sugimoto N (2002) Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur J Biochem 269:2821–2830
Biggar KK, Storey KB (2014) Insight into temperature-dependent microRNA function in mammalian hibernators. Temperature 1:84–86
Biggar KK, Storey KB (2015) Low-temperature microRNA expression in the painted turtle, Chrysemys picta during freezing stress. FEBS Lett 589:3665–3670
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524
Huang Y, Shen XJ, Zou Q et al (2011) Biological functions of microRNAs: a review. J Physiol Biochem 67:129–139
Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10:185–191
Jaskiewicz L, Filipowicz W (2008) Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320:77–97
MacFarlane L-A, Murphy PR (2010) MicroRNA: biogenesis, function and role in cancer. Curr Genom 11:537–561
Lee HY, Doudna JA (2012) TRBP alters human precursor microRNA processing in vitro. RNA 18:2012–2019
Fukunaga R, Han BW, Hung J-H et al (2012) Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151:533–546
O’Carroll D, Schaefer A (2013) General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 38:39–54
Abe M, Bonini NM (2013) MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol 23:30–36
Storey KB, Storey JM (2007) Tribute to PL Lutz: putting life on ‘pause’—molecular regulation of hypometabolism. J Exp Biol 210:1700–1714
Greenway SC, Storey KB (2000) Activation of mitogen-activated protein kinases during natural freezing and thawing in the wood frog. Mol Cell Biochem 209:29–37
Cai Q, Storey KB (1997) Upregulation of a novel gene by freezing exposure in the freeze-tolerant wood frog (Rana sylvatica). Gene 198:305–312
Aguilar OA, Hadj-Moussa H, Storey KB (2016) Regulation of SMAD transcription factors during freezing in the freeze tolerant wood frog, Rana sylvatica. Comp Biochem Physiol B Biochem Mol Biol 201:64–71
Sullivan KJ, Storey KB (2012) Environmental stress responsive expression of the gene li16 in Rana sylvatica, the freeze tolerant wood frog. Cryobiology 64:192–200
Wu S, De Croos JNA, Storey KB (2008) Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica. Gene 424:48–55
Wu S, Storey KB (2005) Up-regulation of acidic ribosomal phosphoprotein P0 in response to freezing or anoxia in the freeze tolerant wood frog, Rana sylvatica. Cryobiology 50:71–82
Dieni CA, Storey KB (2014) Protein kinase C in the wood frog, Rana sylvatica: reassessing the tissue-specific regulation of PKC isozymes during freezing. PeerJ 2:e558
Bansal S, Luu BE, Storey KB (2016) MicroRNA regulation in heart and skeletal muscle over the freeze–thaw cycle in the freeze tolerant wood frog. J Comp Physiol B 186:229–241
Biggar KK, Dubuc A, Storey K (2009) MicroRNA regulation below zero: differential expression of miRNA-21 and miRNA-16 during freezing in wood frogs. Cryobiology 59:317–321
Hadj-Moussa H, Moggridge JA, Luu BE et al (2016) The hibernating South American marsupial, Dromiciops gliroides, displays torpor-sensitive microRNA expression patterns. Sci Rep 6:24627
Chen M, Storey KB (2014) Large-scale identification and comparative analysis of miRNA expression profile in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar Genom 13:39–44
Biggar KK, Kornfeld SF, Maistrovski Y, Storey KB (2012) MicroRNA regulation in extreme environments: differential expression of microRNAs in the intertidal snail Littorina littorea during extended periods of freezing and anoxia. Genom Proteom Bioinform 10:302–309
Lyons PJ, Crapoulet N, Storey KB, Morin PJ (2015) Identification and profiling of miRNAs in the freeze-avoiding gall moth Epiblema scudderiana via next-generation sequencing. Mol Cell Biochem 410:155–163
Lyons PJ, Lang-Ouellette D, Morin PJ (2013) CryomiRs: towards the identification of a cold-associated family of microRNAs. Comp Biochem Physiol Part D Genom Proteom 8:358–364
Tang X, Wen S, Zheng D et al (2013) Acetylation of Drosha on the N-terminus inhibits its degradation by ubiquitination. PLoS One 8:e72503
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016
Okada C, Yamashita E, Lee SJ et al (2009) A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326:1275–1279
Winter J, Jung S, Keller S et al (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Lee Y, Hur I, Park S-Y et al (2006) The role of PACT in the RNA silencing pathway. EMBO J 25:522–532
Wang D, Zhang Z, O’Loughlin E et al (2012) Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26:693–704
Shen J, Xia W, Khotskaya YB et al (2013) EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497:383–387
Godlewski J, Bronisz A, Nowicki MO et al (2010) microRNA-451: a conditional switch controlling glioma cell proliferation and migration. Cell Cycle 9:2742–2748
Tian Y, Nan Y, Han L et al (2012) MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol 40:1105–1112
Yu D, dos Santos CO, Zhao G et al (2010) miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev 24:1620–1633
Xie J, Hu X, Yi C et al (2016) MicroRNA-451 protects against cardiomyocyte anoxia/reoxygenation injury by inhibiting high mobility group box 1 expression. Mol Med Rep 13:5335–5341
Giniatullin AR, Darios F, Shakirzyanova A et al (2006) SNAP25 is a pre-synaptic target for the depressant action of reactive oxygen species on transmitter release. J Neurochem 98:1789–1797
Biggar KK, Wu C-W, Storey KB (2014) High-throughput amplification of mature microRNAs in uncharacterized animal models using polyadenylated RNA and stem–loop reverse transcription polymerase chain reaction. Anal Biochem 462:32–34
Luu BE, Storey KB (2015) Dehydration triggers differential microRNA expression in Xenopus laevis brain. Gene 573:64–69
Costanzo JP, Lee RE, Lortz PH (1993) Physiological responses of freeze-tolerant and -intolerant frogs: clues to evolution of anuran freeze tolerance. Am J Physiol 265:R721–R725
Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475:324–332
Storey KB, Storey JM (2012) Insect cold hardiness: metabolic, gene, and protein adaptation. Can J Zool 90:456–475
Jiang J-J, Xia E-H, Gao C-W, Gao L-Z (2016) The complete mitochondrial genome of western painted turtle, Chrysemys picta bellii (Chrysemys, Emydidae). Mitochondrial DNA 27:787–788
Ramaglia V, Buck LT (2004) Time-dependent expression of heat shock proteins 70 and 90 in tissues of the anoxic western painted turtle. J Exp Biol 207:3775–3784
Storey K, Storey JM (2011) Heat shock proteins and hypometabolism: adaptive strategy for proteome preservation. Res Rep Biol 2:57–68
Naicker MC, Jo IS, Im H (2012) Identification of chaperones in freeze tolerance in Saccharomyces cerevisiae. J Microbiol 50:882–887
Pei W, Tanaka K, Huang SC et al (2016) Extracellular HSP60 triggers tissue regeneration and wound healing by regulating inflammation and cell proliferation. NPJ Regen Med 1:16013
Moon J, Xu L, Giffard RG (2013) Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab 33:1976–1982
Xu L-J, Ouyang Y-B, Xiong X et al (2015) Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp Neurol 264:1–7
Sacconi A, Biagioni F, Canu V et al (2012) miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis 3:e423
Akhtar RS, Ness JM, Roth KA (2004) Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim Biophys Acta Mol Cell Res 1644:189–203
Ma Q, Dasgupta C, Li Y et al (2016) Inhibition of microRNA-210 provides neuroprotection in hypoxic–ischemic brain injury in neonatal rats. Neurobiol Dis 89:202–212
Chung ACK, Huang XR, Meng X, Lan HY (2010) miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21:1317–1325
Hou ST, Nilchi L, Li X et al (2015) Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep 5:7890
Yan-Chun L, Hong-Mei Y, Zhi-Hong C et al (2017) MicroRNA-192-5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Appl Immunohistochem Mol Morphol 25:251–260
Ryan MM, Ryan B, Kyrke-Smith M et al (2012) Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS One 7:e40538
Aguilar OA, Hadj-Moussa H, Storey KB (2017) Freeze-responsive regulation of MEF2 proteins and downstream gene networks in muscles of the wood frog, Rana sylvatica. J Therm Biol 67:1–8
Ye W, Lv Q, Wong C-KA et al (2008) The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One 3:e1719
Szklarczyk D, Franceschini A, Wyder S et al (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Zhang J, Storey KB (2016) RBioplot: an easy-to-use R pipeline for automated statistical analysis and data visualization in molecular biology and biochemistry. PeerJ 4:e2436
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Acknowledgements
This work was supported by a Discovery Grant (Grant #6793) from the Natural Sciences and Engineering Research Council (NSERC) of Canada. KBS holds the Canada Research Chair in Molecular Physiology and HH holds a NSERC Postgraduate scholarship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2018_2821_MOESM1_ESM.xlsx
Primers used for analysis of miRNA expression in the brain of R. sylvatica, including miRNA-specific forward primers, universal reverse primer, and the stem-loop adapter for reverse-transcription. (XLSX 13 kb)
18_2018_2821_MOESM2_ESM.xlsx
Relative expression levels of 113 miRNA species examined in the brain of R. sylvatica. MicroRNA relative expression was evaluated by RT-qPCR of reverse-transcribed, polyadenylated transcripts. Data represent means of n = 3–4 biological replicates from different animals ± SEM. Relative expression of genes was calculated by standardizing against snord68 expression. Control values were adjusted to 1 and the 24 h frozen and 8 h thawed values were expressed relative to the control. Statistical testing used a one-wat ANOVA with a Dunnett's post hoc test, *p < 0.05, **p < 0.01, and ***p <0.001. (XLSX 18 kb)
18_2018_2821_MOESM3_ESM.xlsx
Identity of the significantly enriched biological processes, protein members, and miRNA species identified using MCL clustered protein networks and functional GO ANALYSIS of the miRNAs downregulated in 24 h frozen frog brain. (XLSX 17 kb)
18_2018_2821_MOESM4_ESM.xlsx
Identity of the significantly enriched biological processes, protein members, and miRNA species identified using MCL clustered protein networks and functional GO ANALYSIS of the miRNAs downregulated in 8 h thawed frog brain. (XLSX 22 kb)
18_2018_2821_MOESM5_ESM.jpg
Full cluster map of functional target enrichment and network clustering of the subset of miRNAs downregulated in the brains of 24 h frozen wood frogs. Downstream miRNA target prediction was performed at -2 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S3 for more information on individual clusters, proteins, and the targeting of individual miRNA species. (JPEG 351 kb)
18_2018_2821_MOESM6_ESM.jpg
Full cluster map of functional target enrichment and network clustering of the subset of miRNAs downregulated in the brains of 8 h thawed wood frogs. Downstream miRNA target prediction was performed at 5 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S4 for more information on individual clusters, proteins, and the targeting of individual miRNA species. (JPEG 302 kb)
Rights and permissions
About this article
Cite this article
Hadj-Moussa, H., Storey, K.B. Micromanaging freeze tolerance: the biogenesis and regulation of neuroprotective microRNAs in frozen brains. Cell. Mol. Life Sci. 75, 3635–3647 (2018). https://doi.org/10.1007/s00018-018-2821-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2821-0