Abstract
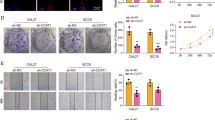
Lysine-specific demethylase 2A (KDM2A, also known as JHDM1A or FBXL11) plays an important role in regulating cell proliferation. However, the mechanisms on KDM2A controlling cell proliferation are varied among cell types, even controversial conclusions have been drawn. In order to elucidate the functions and underlying mechanisms for KDM2A controlling cell proliferation and apoptosis, we screened a KDM2A knockout HEK293T cell lines by CRISPR–Cas9 to illustrate the effects of KDM2A on both biological process. The results indicate that knocking down expression of KDM2A can significantly weaken HEK293T cell proliferation. The cell cycle analysis via flow cytometry demonstrate that knockdown expression of KDM2A will lead more cells arrested at G2/M phase. Through the RNA-seq analysis of the differential expressed genes between KDM2A knockdown HEK293T cells and wild type, we screened out that TGF-β pathway was significantly downregulated in KDM2A knockdown cells, which indicates that TGF-β signaling pathway might be the downstream target of KDM2A to regulate cell proliferation. When the KDM2A knockdown HEK293T cells were transient-transfected with KDM2A overexpression plasmid or treated by TGF-β agonist hydrochloride, the cell proliferation levels can be partial or completely rescued. However, the TGF-β inhibitor LY2109761 can significantly inhibit the KDM2A WT cells proliferation, but not the KDM2A knockdown HEK293T cells. Taken together, these findings suggested that KDM2A might be a key regulator of cell proliferation and cell cycle via impacting TGF-β signaling pathway.







Similar content being viewed by others
References
Klein BJ et al (2018) Recognition of cancer mutations in histone H3K36 by epigenetic writers and readers. Epigenetics 13:683–692
Huang Y et al (2015) Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol 36(1):271–278
Zhuang L et al (2018) Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat Commun 9(1):1796
Li C et al (2018) Histone methyltransferase SETD2 is required for meiotic maturation in mouse oocyte. J Cell Physiol 234:661–668
Faundes V et al (2018) Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am J Hum Genet 102(1):175–187
Dhar SS et al (2014) Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem 289(11):7483–7496
Gao R et al (2013) Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15INK4B and p27Kip1. Mol Cell Biochem 379(1–2):115–122
Kawakami E et al (2015) The histone demethylase Fbxl11/Kdm2a plays an essential role in embryonic development by repressing cell-cycle regulators. Mech Dev 135:31–42
Chen JY et al (2017) Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis 6(8):e369
Lu T et al (2010) Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA 107(1):46–51
Borgel J et al (2017) KDM2A integrates DNA and histone modification signals through a CXXC/PHD module and direct interaction with HP1. Nucleic Acids Res 45(3):1114–1129
Frescas D et al (2008) KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 7(22):3539–3547
Wagner KW et al (2013) KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest 123(12):5231–5246
Rizwani W et al (2014) Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS ONE 9(7):e100888
Tanaka Y et al (2015) Mild Glucose starvation induces KDM2A-mediated H3K36me2 demethylation through AMPK to reduce rRNA transcription and cell proliferation. Mol Cell Biol 35(24):4170–4184
Du J et al (2013) Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells 31(1):126–136
Yu G et al (2016) Demethylation of SFRP2 by histone demethylase KDM2A regulated osteo-/dentinogenic differentiation of stem cells of the apical papilla. Cell Prolif 49(3):330–340
Yang M et al (2018) TNFAIP3 is required for FGFR1 activation-promoted proliferation and tumorigenesis of premalignant DCIS.COM human mammary epithelial cells. Breast Cancer Res 20(1):97
Gunaratne A et al (2015) aPKC alters the TGFbeta response in NSCLC cells through both Smad-dependent and Smad-independent pathways. J Cell Sci 128(3):487–498
Wei Y et al (2018) Epigenetic modifications in KDM lysine demethylases associate with survival of early-stage NSCLC. Clin Epigenetics 10:41
Kong Y et al (2016) RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett 381(1):138–148
Cao LL et al (2018) Lysine-specific demethylase 2A expression is associated with cell growth and cyclin D1 expression in colorectal adenocarcinoma. Int J Biol Markers https://doi.org/10.1177/1724600818764069
Liu L et al (2018) Activation of peroxisome proliferation-activated receptor-gamma inhibits transforming growth factor-beta1-induced airway smooth muscle cell proliferation by suppressing Smad-miR-21 signaling. J Cell Physiol 234:669–681
Yan X et al (2018) CXXC5 suppresses hepatocellular carcinoma by promoting TGF-beta-induced cell cycle arrest and apoptosis. J Mol Cell Biol 10(1):48–59
Tanaka Y et al (2010) JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J 29(9):1510–1522
Lu DH et al (2018) Lysine demethylase 2A promotes the progression of ovarian cancer by regulating the PI3K pathway and reversing epithelial mesenchymal transition. Oncol Rep 41:917–927
Acknowledgements
This study was supported by the National Natural Science Foundation of China (30900816 and 31371307) and the Fundamental Research Funds for the South-Central University for Nationalities (CZP17082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests for this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, Wh., Liang, Dy., Wang, Q. et al. Knockdown of KDM2A inhibits proliferation associated with TGF-β expression in HEK293T cell. Mol Cell Biochem 456, 95–104 (2019). https://doi.org/10.1007/s11010-018-03493-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-018-03493-5