Abstract
Null hypothesis significance testing is routinely used for comparing the performance of machine learning algorithms. Here, we provide a detailed account of the major underrated problems that this common practice entails. For example, omnibus tests, such as the widely used Friedman test, are not appropriate for the comparison of multiple classifiers over diverse data sets. In contrast to the view that significance tests are essential to a sound and objective interpretation of classification results, our study suggests that no such tests are needed. Instead, greater emphasis should be placed on the magnitude of the performance difference and the investigator’s informed judgment. As an effective tool for this purpose, we propose confidence curves, which depict nested confidence intervals at all levels for the performance difference. These curves enable us to assess the compatibility of an infinite number of null hypotheses with the experimental results. We benchmarked several classifiers on multiple data sets and analyzed the results with both significance tests and confidence curves. Our conclusion is that confidence curves effectively summarize the key information needed for a meaningful interpretation of classification results while avoiding the intrinsic pitfalls of significance tests.
Similar content being viewed by others
1 Introduction
Machine learning classifiers are frequently compared and selected based on their performance on multiple benchmark data sets. Given a set of k classifiers and N data sets, the question is whether there exists a significant performance difference, and if so, between which pairs of classifiers. Null hypothesis significance testing (NHST) is increasingly used for this task. However, NHST has been criticized for many years in other fields (Harlow et al. 1997), for example, biomedicine and epidemiology (Poole 1987; Goodman 1993, 2008; Rothman et al. 2008; Stang et al. 2010), the social sciences (Cohen 1994; Gigerenzer et al. 2004), statistics (Berger and Berry 1988), and particularly psychology (Rozeboom 1960; Bakan 1966; Carver 1978; Schmidt and Hunter 1997; Rozeboom 1997; Gigerenzer 1998). By contrast, in machine learning, these critical voices have not been widely echoed so far. Recently, some deficiencies of the common benchmarking practice have been pointed out (Drummond and Japkowicz 2010), and Bayesian alternatives were proposed (Corani et al. 2015; Benavoli et al. 2015). Overall, however, there is a clear trend towards significance testing for the comparison of machine learning algorithms.
In this paper, we criticize this common evaluation practice. First, we scrutinize the key problems and common misconceptions of NHST that, in our view, have received scant attention in the machine learning literature so far. For example, it is widely assumed that NHST originates from one coherent theory (Goodman 2008), but actually it is an unfortunate hybrid of concepts from the Fisherian and Neyman–Pearsonian school of thought. We believe that the amalgamation of incompatible ideas from these schools and the ensuing problems are not widely recognized. For example, the p value is often considered a type of error rate, although it does not have such an interpretation. A p value is widely considered as an objective measure, but in fact, it depends on the researcher’s intentions (whether these were actually realized or not) and how the researcher thought about the experiment. The p value is therefore far less objective than is commonly assumed. Sampling intentions do matter, and they also have a bearing on other frequentist methods, such as confidence intervals.
A significant p value is widely regarded as a research desideratum, but it is probably one of the most widely misinterpreted and overrated values in the scientific literature (Goodman 2008; Nuzzo 2014). We investigate several problems of this recondite value with particular relevance to performance evaluation. A major goal of this study is to kindle a debate on the role of NHST, the p value, and alternative evaluation methods in machine learning.
One of our main criticisms concerns the use of omnibus tests in comparative classification studies. The Friedman test is now widely used when multiple classifiers are compared over multiple data sets. When such tests give a significant result, post-hoc tests are carried out to detect which pair-wise comparisons are significantly different. Here, we provide several arguments against this procedure in general and the Friedman test in particular. A key finding is that such tests are not needed, and when a study involves diverse benchmark data sets, omnibus tests (such as the Friedman test) are not even appropriate.
The underlying problem of the current evaluation practice, however, is a much deeper one. There is a common thread that weaves through the machine learning literature, suggesting that statistical testing lends scientific rigor to the analysis of empirical results. Well-meaning researchers, eager for a sound and objective interpretation of their empirical results, might consider a statistical test indispensable. Here, we wish to challenge this view. We argue that such tests often provide only a veneer of rigor, and that they are therefore not needed for the comparison of classifiers. Our criticism pertains to both the Fisherian significance testing and the Neyman–Pearsonian hypothesis testing, and particularly to the blurring of concepts from both schools of thought. We do not consider Bayesian testing in this article.
We put forward that a focus on the effect size (i.e., the magnitude of the difference in performance) and its reasonable bounds is needed, not a focus on statistical significance. As an alternative evaluation tool, we propose confidence curves, which are based on the idea of depicting an infinite number of nested confidence intervals for an effect size (Birnbaum 1961). The resulting “tipi”-shaped graph enables the investigator to simultaneously assess the compatibility of an infinite number of null hypotheses with the experimental results. Thereby, confidence curves solve a key problem of the common testing practice, namely the focus on a single null hypothesis (i.e., the null hypothesis of no difference) with its single p value.
In our experiments involving real-world and synthetic data sets, we use first the Friedman test with Nemenyi post-hoc test and then confidence curves. By juxtaposing both approaches, we show that the evaluation with confidence curves is more meaningful but, at the same time, also more challenging because they require an interpretation beyond the dichotomous decision of “significant” versus “non-significant”.
The novelty of this study is twofold. First, we investigate several underrated problems of NHST and the p value. To our knowledge, no detailed account of these problems has been given in the machine learning literature yet. Second, we propose confidence curves as an alternative, graphical evaluation tool. The significance of our work is that it opens a possible avenue towards a more flexible and meaningful interpretation of empirical classification results. The main contributions of our paper are as follows.
-
We investigate five key problems of the p value that are particularly relevant for the evaluation of classification results but have received scant attention so far. We discuss several examples to illustrate these problems.
-
We show that widely used omnibus tests, such as the Friedman test, are not appropriate for the comparison of multiple classifiers over multiple data sets. If the test subjects are diverse benchmark data sets, then the p value has no meaningful interpretation.
-
We propose an alternative evaluation method, confidence curves, which help avoid the intrinsic pitfalls of NHST. As a summary measure, we derive the area under the confidence curve (AUCC). We provide a detailed experimental comparison between the evaluation based on NHST and confidence curves.
-
We provide the R code to plot confidence curves and calculate the AUCC. This code is available at https://github.com/dberrar/ConfidenceCurve.
This paper is organized as follows. After a brief review of related work, we first describe the main differences between the Fisherian and the Neyman–Pearsonian school of thought, which are often amalgamated into an incoherent framework for statistical inference. Then, we scrutinize the key problems of the p value. Finally, we present several arguments against significance tests for the comparison of multiple classifiers over multiple data sets. This first part of the paper represents the rationale for our research on alternative evaluation methods. We begin the second part of the paper with an illustration of the key concepts of confidence curves and then provide their mathematical details. As a summary statistic of precision, we propose the area under the confidence curve (AUCC). Then, we provide some examples illustrating what we can do with these curves and the AUCC. In the experimental part of the paper, we compare the performance of several classifiers over both UCI benchmark and synthetic data sets. First, we analyze the results using a standard approach (Friedman test with Nemenyi post-hoc test). Then, we interpret the same results with confidence curves and compare both approaches. In Sect. 8, we summarize our arguments against significance testing and discuss the pros and cons of the proposed alternative. Our conclusion (Sect. 9) is that greater emphasis should be placed on effect size estimation and informed judgment, not on significance tests and p values.
2 Related work
There exists a substantial amount of literature on the problems of significance testing. In machine learning, however, such critical voices are extremely rare. Demšar (2006), for example, concludes with a paragraph reminding us about the alternative opinion of statisticians who reject statistical testing (Cohen 1994; Schmidt 1996). These objections are further expatiated in (Demšar 2008). Dietterich (1998) compared several statistical tests and concluded that they should be viewed as approximate, heuristic tests, and not as rigorously correct statistical methods. Drummond and Japkowicz (2010) criticize the current practice in machine learning that puts too much emphasis on benchmarking and statistical hypothesis testing. In a similar vein, Drummond (2006) questions the value of NHST for comparing the performance of machine learning algorithms.
Yet despite decades of severe criticisms, significance tests and their p values enjoy an unbroken popularity in many scientific disciplines (Nuzzo 2014). How prevalent is their use in machine learning? As it is difficult to answer that question directly, we queried the ScienceDirect databaseFootnote 1 for articles containing the terms “p value” and “classification”. We divided the number of articles containing these search terms by the number of articles containing only “classification” and not “p value”. We restricted the search to computer science articles only. Figure 1 shows that p values (and hence significance testing) have been increasingly used over the last 15 years. Of course, Fig. 1 needs to be interpreted very cautiously because the results may also include articles that are critical of significance testing. Nonetheless, we believe that Fig. 1 indicates a clear trend towards the use of significance tests for the comparison of classifiers.
Use of significance tests in classification studies. The rate denotes the number of computer science articles containing the words “p value” and “classification” divided by the number of computer science articles containing only “classification” and not “p value”. Results are based on ScienceDirect database queries, 19 December 2015
Several alternatives to significance testing have been proposed, for example, Bayesian analysis (Berger and Berry 1988). Bayesian tests were also recently proposed for the comparison of machine learning algorithms (Benavoli et al. 2015; Corani et al. 2015). Killeen (2004) recommends replacing the p value by \(p_{rep}\), a measure of replicability of results.
As another alternative, confidence intervals are widely considered more meaningful than significance tests (Tukey 1991; Cohen 1994; Schmidt 1996). In fact, a confidence interval provides a measure of the effect size and a measure of its uncertainty (Cummings 2012), whereas the p value conflates the effect size with the precision with which this effect size has been measured. We will discuss this issue in detail in Sect. 4.2. Many statisticians and other scientists have therefore argued that confidence intervals should replace significance tests and p values (Cox 1977; Cohen 1994; Schmidt 1996; Thompson 1999; Stang et al. 2010). The journal Epidemiology even advises against the use of p values: “[...] we prefer that p values be omitted altogether, provided that point and interval estimates, or some equivalent, are available.” (Rothman 1998, p. 334). Although confidence intervals and p values are often considered as two sides of the same coin, they are different tools and have a different influence on the interpretation of empirical results (Poole 2001). Specifically, for the comparison of machine learning classifiers, confidence intervals were shown to be preferable to significance tests (Berrar and Lozano 2013).
However, Levin (1998), Savalei and Dunn (2015), and Abelson (1997), among others, are skeptical about the benefits of confidence intervals over significance testing because it is unclear how wide such intervals should be. It has been suggested that several intervals alongside the common 95% interval be reported (Cox 1958), but according to Levin (1998), this is “subjective nonsense” (p. 47) because it is unclear (and arbitrary) which confidence levels should be reported. Furthermore, it is perhaps too tempting to interpret a confidence interval merely as a surrogate significance test by checking whether it includes the null value or not. In that case, the advantage of the confidence interval over the p value is of course lost.
The evaluation method that we consider as an alternative to NHST is based on the confidence curve estimator developed by Birnbaum (1961). In his unified theory of estimation for one-parameter problems, Birnbaum constructed nested confidence intervals for point estimates. He did not propose confidence curve estimators as an alternative to significance testing, though. In epidemiology and medical research, these estimators were proposed as a meaningful inferential tool under their alias of p value function (Poole 1987; Rothman 1998; Rothman et al. 2008). Similar graphs were proposed before under the different names of consonance function (Folks 1981) and confidence interval function (Sullivan and Foster 1990). To our knowledge, however, such graphs are rarely used in epidemiology or clinical research. In reference to the paper that first described the key idea, we use Birnbaum’s term confidence curve to refer to nested confidence intervals at all levels. We consider confidence curves for cross-validated point estimates of classification performance. We also derive the area under the confidence curve (AUCC), which, similarly to the AUC of a ROC curve, is a scalar summary measure.
3 Short revision of classic statistical testing
The foundations of what has become the classic statistical testing procedure were laid in the early 20th century by two different approaches to statistical inference, the Fisherian and the Neyman–Pearsonian school of thought. These two schools are widely believed to represent one single, coherent theory of statistical inference (Hubbard 2004). However, their underlying philosophies and concepts are fundamentally different (Goodman 1993; Hubbard and Bayarri 2003; Hubbard and Armstrong 2006) and their amalgamation can entail severe problems. We will now briefly revise the essential concepts.
3.1 Fisherian significance testing
The Fisherian school of thought goes back to Ronald A. Fisher and is motivated by inductive inference, which is based on the premise that it is possible to make inferences from observations to a hypothesis. In the Fisherian paradigm, only one hypothesis exists, the null hypothesis. There is no alternative hypothesis. Following the notation by Bayarri and Berger (2000), we state the null hypothesis, H0, as follows,
where X denotes data, and \(f({\mathbf{x}},\theta )\) is a density with parameter \(\theta \). The word “null” in “null hypothesis” refers to the hypothesis to be nullified. It does not mean that we need to test whether some value is 0 (for example, that the difference in performance is 0). To make this distinction clear, Cohen (1994) prefers “nil hypothesis” for the null hypothesis of no difference.
In the Fisherian inductive paradigm, we are only interested in whether the null hypothesis is plausible or not. So we ask: which data cast as much doubt as (or more doubt than) our observed data, given that the null hypothesis is true? Fisher considered this conditional probability, called the p value, as a measure of evidence against the null hypothesis: the smaller this value, the greater the evidential weight against the null hypothesis, and vice versa. To investigate the compatibility of the null hypothesis with our observed data \({\mathbf{x}_{\mathrm{obs}}}\), we choose a statistic \(T = t({\mathbf{X}})\), for example, the mean. The p value is defined as
In other words, the p value is the probability of a result as extreme as or more extreme than the observed result, given that the null hypothesis is true. As the null hypothesis is a statement about a hypothetical infinite population, the p value is a measure that refers to that population. The p value is therefore not a summary measure of the observed data at hand.
Under the null hypothesis, the p value is a random variable uniformly distributed over [0, 1]. If the p value is smaller than an arbitrary threshold (commonly 0.05, the Fisherian level of significance), then the result is considered “significant”, otherwise “non-significant”. For Fisher, a significant p value merely meant that it is worthwhile doing further experiments (Goodman 2008; Nuzzo 2014). Formally, the p value is defined as a probability, but it is a rather difficult-to-interpret probability—it may be best to think of the p value as a “crude indicator that something surprising is going on” (Berger and Delampaday 1987, p. 329). As Fisher reminded us, this “something surprising” may also refer to a problem with the study design or the data collection process (Fisher 1943).
We are often reminded not to interpret the p value as the probability that the null hypothesis is true, given the data, that is, Pr(H0\(|{\mathbf{x}_{\mathrm{obs}}}\)). Still, this interpretation is perhaps one of the most pervasive misconceptions of the p value. Sometimes we are advised that although the p value cannot tell us anything about the probability that the null hypothesis is true or false, we can act as if it were true or false. However, this is not in the spirit of Fisher who regarded the p value as an evidential measure, not as a criterion for decision making or behavior. It is the Neyman–Pearsonian hypothesis testing that provides such a criterion.
3.2 Neyman–Pearsonian hypothesis testing
In contrast to the Fisherian paradigm, the procedure invented by Jerzy Neyman and Egon Pearson regards hypothesis testing as a vehicle for decision making or inductive behavior. Here, the null hypothesis H0 is pitted against an alternative hypothesis, H1 (which, we remember, does not exist in the Fisherian paradigm). The emphasis is on making a decision between two options, with the goal to minimize the errors that we make in the long run, not to find out which hypothesis is true. Thus, “accepting H” is not to be equated with “believing that H is true”. In the words of Neyman and Pearson,
We are inclined to think that as far as a particular hypothesis is concerned, no test based upon the theory of probability can by itself provide any valuable evidence of the truth or falsehood of that hypothesis [...] Without hoping to know whether each separate hypothesis is true or false, we may search for rules to govern our behavior with regard to them, in following which we insure that, in the long run of experience, we shall not be too often wrong. (Neyman and Pearson 1933, p. 290–291) (our italics).
The importance of the phrase “in the long run” cannot be overstated. It means that the Neyman–Pearsonian paradigm is not conceived as a procedure to assess the evidential weight provided by an individual experimental outcome.
There are now two errors that one can make, (i) deciding against H0 although it is correct (Type I error or \(\alpha \)), and (ii) deciding in favor of H0 although it is false (Type II error or \(\beta \)). Note that “deciding in favor of” and “deciding against” implies a dichotomization of the results; otherwise, the concept of Type I/II errors would have no meaning. If H0 is false, then the probability—over many applications of the testing procedure—that H0 is rejected is the power of that procedure, where \(\textit{power} = 1 - \beta \). There is a trade-off between the two types of errors, and we can tweak them through our arbitrarily fixed \(\alpha \). What should guide this tweaking? Clearly, the guide should be the costs associated with the errors. Costs, however, have no bearing on the truth of a hypothesis; they have purely pragmatic reasons and were frowned upon by Fisher (1955).
Note that in the Neyman–Pearsonian school of thought, the Fisherian p value does not exist. Nor does any other measure of evidence. By contrast, the concepts of error rates, alternative hypothesis, and power do not exist in the Fisherian school of thought. Specifically, note the crucially important difference between the p value and the Type I error rate. The p value has no interpretation as a long-run, repetitive error rate, whereas the Type I error rate does; compare (Berger and Delampaday 1987, p. 329). This stands in stark contrast to the nearly ubiquitous misinterpretation of p values as error rates. It means that we cannot simply compare the Fisherian p value with the Neyman–Pearsonian error rate \(\alpha \). If the question of interest is “Given these data, do I have reason to conclude that H0 is false?”, then the Neyman–Pearsonian error rate is irrelevant. Note that a Bayesian calibration can, to some extent, reconcile the Fisherian p value and the Neyman–Pearsonian \(\alpha \). The calibration is \(\alpha (p) = [1 + (-ep\log p)^{-1} ]^{-1}\), if \(p<e^{-1}\) (Sellke et al. 2001). For example, if the p value is 0.03, then it can be given the frequentist error interpretation of \(\alpha = 0.22\). A lower bound on the Bayes factor is given by \(B(p) = -ep\log (p)\). For example, a p value of 0.03 corresponds to an odds of 0.29 for H0 to H1 (i.e., about 1:3.5). This calibration also illustrates that a p value almost always drastically overstates the evidence against the null hypothesis.
In the Fisherian school of thought, we can never accept H0, only fail to reject it, if we deem the p value too high. But in the Neyman–Pearsonian paradigm, we may indeed accept H0 because we are interested in decision rules: if the test statistic falls into the rejection region, then H0 is rejected and H1 is accepted. By contrast, if the test statistic does not fall into the rejection region, then H0 is accepted and H1 is rejected. The concrete numerical value of the probability associated with the test statistic is irrelevant. In fact, the Neyman–Pearsonian hypothesis test is based on the notion of a critical region that minimizes \(\beta \) for a fixed \(\alpha \). The concept of “error rate” requires that a result can be anywhere within the tail area (Goodman 1993). This is not so for the p value, and it makes therefore sense to report it exactly. There is no big difference between a p value of 0.048 and 0.052, simply because they can be interpreted as indicators of about equal weight. But in the Neyman–Pearsonian school of thought, it is an all-or-nothing decision, so 0.048 and 0.052 make all the difference.
Confused? You should be. Both the Neyman–Pearsonian \(\alpha \)-level and the Fisherian p value have been called the “significance level of a test”. The \(\alpha \)-level is set before the experiment is carried out, whereas the p value is calculated from the data after the experiment. The symbol \(\alpha \) is used as both an arbitrary threshold for the p value and as a frequentist error rate. And to make matters worse, the two different concepts are commonly employed at the 5%-level, thereby blurring the differences even more. In an excellent review, Hubbard (2004) describes the widespread confusion over p values as error probabilities, which has been perpetuated even in statistics textbooks. This confusion has also percolated into the machine learning literature where we observe the misconception that replicability or power (a Neyman–Pearsonian concept) can be measured as a function of p values (Demšar 2006). The p value, however, tells us nothing about either replicability or power. We will come back to this issue in Sect. 4.4.
While Neyman believed that null hypothesis testing can be “worse than useless” (Gigerenzer 1998, p. 200) in a mathematical sense, Fisher called the Neyman–Pearsonian Type II error the result of a “mental confusion” (Fisher 1955, p. 73). Therefore, Fisher, Neyman, and Pearson would undoubtedly all have strongly objected to the inconsistent conflation of their ideas.
While the Neyman–Pearsonian approach is certainly useful in an industrial quality-control setting—a fact that was acknowledged even by one of its sternest opponents, R. A. Fisher (1955)—it can be questioned whether it has any role to play in the scientific enterprise. Rothman et al. (2008) wonder:
Why has such an unsound practice as the Neyman-Pearson (dichotomous) hypothesis testing become so ingrained in scientific research? [...] The neatness of an apparent clear-cut result may appear more gratifying to investigators, editors, and readers than a finding that cannot be immediately pigeonholed. (Rothman et al. 2008, p. 154)
When we are interested in evaluating the plausibility of a concrete hypothesis, we might ask: “How compatible are our data with the hypothesis?” It seems that the Fisherian significance testing with its (allegedly evidential) p value is indeed more suitable to answer this question than the Neyman–Pearsonian approach. However, the p value is not as easy to interpret as we might think.
4 Underrated problems of the p value
Goodman (2008) gives an overview of the 12 most common misconceptions of the p value. Perhaps the single most serious misconception is that the p value has a sound theoretical foundation as an inferential tool. In fact, Fisher regarded the p value as an evidential measure of the discrepancy between the data and the null hypothesis, which should be used together with other background information to draw conclusions from experiments (Goodman 1999). The p value, however, is not an evidential measure (Berger and Sellke 1987; Goodman and Royall 1988; Cohen 1994; Hubbard and Lindsay 2008; Schmidt and Hunter 1997; Schervish 1996). An evidential measure requires two competing explanations for an observation (Goodman and Royall 1988), but the theory underlying the p value does not allow any alternative hypothesis. The p value is based on only one hypothesis. But at least, the p value is an objective measure—or is it not?
4.1 The p value is not a completely objective measure
The p value includes probabilities for data that were actually not obtained, but that could have been obtained under the null hypothesis. Yet what exactly are these imaginary, more extreme data? Curiously, this question cannot be answered on the basis of the observed data alone, but we need to know how the researcher thought about the possible outcomes. This means that it is impossible to analyze any experimental outcomes for their (non-)significance, unless we understand how the experiment was planned and conducted. We illustrate this problem in two scenarios that are adapted from mathematically equivalent examples of hypothetical clinical trials (Goodman 1999; Berger and Berry 1988).
Suppose that Alice and Bob jointly developed a new classifier A. They believe that A can outperform another classifier X on a range of data sets. Both Alice and Bob formulate the null hypothesis as follows, H0: the probability that their algorithm is better than X is 0.5. Alice and Bob decide to benchmark their algorithm independently and then compare their results later. Both Alice and Bob select the same six data sets from the UCI repository.
Alice is determined to carry out all six benchmark experiments, even if her algorithm loses the competition on the very first few data sets. After all six experiments, she notes that their classifier A was better on the first five data sets, but not on the last data set. Under H0, the probability of observing these results is calculated as \({6 \atopwithdelims ()5}0.5^10.5^5\) (i.e., 5 successes out of 6 trials, where each trial has a chance of 0.5). A more extreme result would be that their algorithm performs better on all six data sets. Thus, the probability of the more extreme result is \(0.5^6\). Therefore, Alice obtains \({6 \atopwithdelims ()5}0.5^10.5^5 + 0.5^6 = 0.11\) as the one-sided p value. Consequently, she concludes that their model A is not significantly better than the competing model X.
Bob has a different plan. He decides to stop the benchmarking as soon as their algorithm fails to be superior. Coincidentally, Bob analyzed the data sets in the same order as Alice did. Consequently, Bob obtained the identical benchmark results. But Bob’s calculation of the p value is different from Alice’s. In Bob’s experiment, the failure can only happen at the end of a sequence of experiments because he planned to stop the benchmarking in case that A performs worse than X. Hence, the probability of the observed result is \(0.5^5\times 0.5^1\) (i.e., a success in the first five experiments and a failure in the last one). The probability of the more extreme result is the same as that that Alice calculated. Therefore, Bob obtains \(0.5^5\times 0.5^1 + 0.5^6 = 0.03\) as the one-sided p value. And he concludes that their classifier is significantly better than the competing model X. The conundrum is that both Bob and Alice planned their experiments well. They used the same data sets and the same models, yet their p values—and in this example, their conclusions—are quite different.
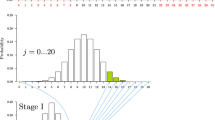
Before Bob continues with his research, he discusses his experimental design with his supervisor, Carlos. Bob plans to compare their algorithm with an established algorithm X. This time, the null hypothesis is stated as follows, H0: probability that his algorithm performs differently from X is 0.5. Thus, this time, it is a two-sided test. Bob decides to benchmark his algorithm against X on ten data sets. He observes that his algorithm outperforms X in 9 of 10 data sets. The probability of 9 successes in 10 trials is \({10 \atopwithdelims ()9}0.5^90.5^1=0.0098\). The possible outcomes that are as extreme as or more extreme than the observed outcome are 0, 1, 9, and 10. The two-sided p value is the sum of probabilities of these outcomes: \({10 \atopwithdelims ()0} 0.5^0 0.5^{10} + {10 \atopwithdelims ()1} 0.5^1 0.5^{9} + {10 \atopwithdelims ()9} 0.5^9 0.5^{1} + {10 \atopwithdelims ()10} 0.5^{10} 0.5^{0}= 0.021\). Given that the p value is smaller than 0.05, Bob concludes that his algorithm is significantly better than X.
Alice discusses a different study design with Carlos. She also wants to investigate the same ten data sets; however, if she cannot find a significant difference after ten experiments, then she wants to investigate another ten data sets. Thus, her study has two stages, where the second stage is merely a contingency plan. Her null hypothesis is the same as Bob’s. She uses again the same data sets as Bob did, and in the same order. Therefore, she also observes that her algorithm outperforms X in 9 out of 10 data sets. Thus, she does not need her contingency plan.
However, after discussing her result with Carlos, she is disappointed: her two-sided p value is 0.34. How can that be? After all, she did exactly the same experiments as Bob did and obtained exactly the same results! This is highly counterintuitive, but it follows from the logic of the p value. After the first 10 experiments, the outcomes that are as extreme as her actually observed results are 1 and 9. The more extreme results are 0 and 10. In the two-stage design, however, the total number of possible experiments is 20. Thus, the two-sided p value is calculated as \({20 \atopwithdelims ()0}0.5^0 0.5^{20} + {20 \atopwithdelims ()1}0.5^1 0.5^{19} + {20 \atopwithdelims ()9}0.5^9 0.5^{11} + {20 \atopwithdelims ()10}0.5^{10} 0.5^{10} = 0.34\). In the words of Berger and Berry (1988), “[...] the nature of p values demands consideration of any intentions, realized or not” (p. 165). It does not matter that Alice did not carry out her contingency plan. What matters is that she contemplated to do so before obtaining her results, and this does affect the calculation of her p value.
Even if sufficient evidence against a hypothesis has been accrued, the experiment must adhere to the initial design; otherwise, p values have no valid interpretation. This is the reason why clinical trials cannot be stopped in mid-course for an interim (frequentist) analysis. In practice, it can happen that a drug trial is stopped in mid-term because the beneficial effects of the drug are so clear that it would be unethical to continue administering a placebo to the control group (Morgan 2003). But then the data can be analyzed only by Bayesian, not frequentist methods. The frequentist paradigm requires that the investigator adhere to everything that has been specified before the study (Berry 2006). Realized or not, the intentions of the investigator matter, which indicates a potentially serious flaw in the logic of the p value (Berger and Berry 1988).
4.2 The p value conflates effect size and precision
A common misinterpretation of the p value is that it reflects the probability that the experiment would show as strong an effect as the observed one (or stronger), if the null hypothesis was correct. Suppose that we observe a difference in accuracy of 0.15 between two classifiers, A and B, with an estimated standard deviation of 0.10. For simplicity, let us assume that a standard normal test statistic is appropriate. Then we obtain \(z = \frac{0.15}{0.10}=1.5\) with a two-tailed p value of \(0.134 > 0.05\). Thus, we cannot reject the null hypothesis of equal performance between A and B. Now assume that we observe a difference of only 0.12 between A and C, with an estimated standard deviation of 0.06. The test statistic \(z = \frac{0.12}{0.06}=2\) gives a p value of 0.046. Thus, we can reject the null hypothesis of equal performance between A and C. But compared with the difference between A and B, the difference between A and C is smaller (and closer to the null value of 0). The problem is that the p value conflates the magnitude of the difference (here, 0.15 and 0.12, respectively) with its precision (measured by the standard deviations 0.10 and 0.06, respectively).
To illustrate the conflating effect, we compared the performance of random forest and CART on the data set Transfusion from the UCI repository in r times repeated tenfold stratified cross-validation (Fig. 2). The p values were derived from the variance-corrected t test (Nadeau and Bengio 2003; Bouckaert and Frank 2004).Footnote 2 The test statistic,
follows approximately Student’s distribution with \(\nu =kr-1\) degrees of freedom; \(a_{ij}\) and \(b_{ij}\) denote the performances (here, accuracy) achieved by classifiers A and B, respectively, in the jth repetition of the ith cross-validation fold; s is the standard deviation; \(n_2\) is the number of cases in one validation set, and \(n_1\) is the number of cases in the corresponding training set.
For \(r=1\), i.e., a single run of tenfold cross-validation, the p value is 0.065, so we do not see a significant difference between the two classifiers. However, by repeating the cross-validation just three times (\(r=3\)), the p value falls below the magical 5% hurdle. The p value decreases further with increasing r. For 10 times repeated tenfold cross-validation, we obtain a p value of 0.003.
It is a well-known fact that for a large enough sample size, we are bound to find a statistically significant result (Hays 1963). With an increasing number of repetitions, we can increase the sample size. Note, however, that the differences \(a_{ij}-b_{ij}\) become more and more dependent, since we are effectively analyzing the same data over and over again. This is of course a clear violation of the assumptions of the test; however, this violation is not immediately obvious. Depending on our intentions, we could now choose to present either the result of the single cross-validation (“the observed difference was not significant”) or the result of the 10 times repeated tenfold cross-validation (“the observed difference was highly significant”).
4.3 Same p value, different null hypothesis
It is not generally appreciated that for every point null hypothesis, there is another point null hypothesis, possibly with a very different null value, that has exactly the same p value. We call it the entangled null hypothesis. Consider the following example. Bob and Alice analyze independently the accuracy of two classifiers, A and X, where X is an established classifier and A is their novel classifier. Bob’s null hypothesis is that there is no difference in performance, i.e., \(\text {H0:}~\delta = \tau _A - \tau _X = 0\), where \(\tau _A\) and \(\tau _X\) refer to the true, unknown accuracy of classifier A and X, respectively.
Alice considers another null hypothesis, \(\text {H0:}~\delta = \tau _A - \tau _X = 0.10\). Suppose now that both Alice and Bob obtain exactly the same p value of, say, 0.15. Bob does not reject the hypothesis of equal performance. Alice, on the other hand, concludes that the data are consistent with a rather large difference (10%) in performance. Thus, neither the null hypothesis of no difference nor its entangled hypothesis of a relatively large difference can be rejected, since the data are consistent with both hypotheses. These results cannot be easily reconciled within the framework of significance testing; with confidence curves, however, they can.
4.4 p value and replicability
Let us consider the following example. Suppose that Alice carried out an experiment \(E_{a1}\) and obtained a p value of \(p_{a1}=0.001\). Bob carried out another experiment, \(E_{b1}\), and obtained \(p_{b1}=0.03\). Now, Alice is going to repeat her experiment. In her new experiment, \(E_{a2}\), everything is the same as in \(E_{a1}\). The only difference is that Alice is going to use a different sample. The size of this sample will be the same as that of \(E_{a1}\). For example, the sample from \(E_{a1}\) is a test set of \(n_{a1}=100\) cases, and the sample from \(E_{a2}\) is a new test set of \(n_{a2}=100\) new cases, which are randomly drawn from the same population of interest. Bob, too, repeats his experiment in this way. We invite the reader to ponder briefly over the following question: who is more likely to get a significant result in the second experiment, Alice (who obtained \(p_{a1}=0.001\)) or Bob (who obtained \(p_{b1}=0.03\))?
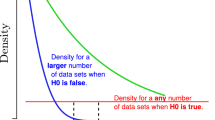
One might be tempted to answer “Alice—because her initial p value is much smaller than Bob’s. Surely, the p value must tell us something about the replicability of a finding, right?” But this is not so. Our interpretation of replicability implies that the smaller the first p value is, the more likely it is that the second p value (from an exact replication study) will be smaller than 0.05, given that the null hypothesis is false. Under the null hypothesis, the p value takes on values randomly and uniformly between 0 and 1. But if the null hypothesis is false, it is the statistical power that determines whether a result can be replicated or not. Power depends on three main factors, (i) the alpha level for the test (the higher the level, the higher the power of the test, everything else being equal); (ii) the true effect size in the target distribution (the larger this effect, the higher the power of the test, everything else being equal); and (iii) the sample size (the larger the test set, the higher the power of the test, everything else being equal) (Fraley and Marks 2007; Schmidt and Hunter 1997). Note that these factors are constants. If the true difference in performance is \(\delta \), and the alpha level is fixed, and the size of the test set is n, then the power of our test is the same in any study, regardless of the concrete makeup of the sampled test set. By contrast, the p value does depend on the concrete makeup. The power of a test to detect a particular effect size in the population of interest can be calculated before the experiment has been carried out. By contrast, the p value can be calculated only after the experiment has been carried out. As the power does not depend on the p value, the p value is irrelevant for assessing the likelihood of replicability. We remember that power is a concept from the Neyman–Pearsonian school of thought, while the p value is a concept from the Fisherian school of thought. Carver (1978) notes:
It is a fantasy to hold that statistical significance reflects the degree of confidence in the replicability or reliability of results. (Carver 1978, p. 384)
The misinterpretation of a significant result as an indicator of replicability is known as replication fallacy.
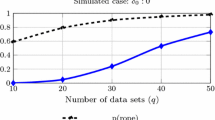
Greenwald et al. (1996) showed that the p value is monotonically related to the replicability of a non-null finding. However, in their study, replicability is understood differently, namely as the probability of data at least as extreme as the observed data under an alternative hypothesis H1, which is defined post-hoc and with the effect size from the first experiment, i.e., Pr (observed or more extreme data|H1). Numerical examples can be found in (Krueger 2001).
4.5 The p value and Jeffreys–Lindley paradox
Suppose that we compare four classifiers over 50 data sets and use a significance test for the null hypothesis of equal performance. Assume that we obtain a very small p value of, say, 0.005. Many researchers might think that it is now straightforward how to interpret this result; however, it is actually not so obvious. The reason is the Jeffreys–Lindley paradox. This paradox is a well-known conundrum in inferential statistics where the frequentist and Bayesian approach give different results. Assume that H0 is a point null hypothesis and x the result of an experiment. Then the following two statements can be true simultaneously (Lindley 1957):
-
1.
A significance test reveals that x is significant at level \(\alpha \).
-
2.
The posterior probability for H0 given the result x, \(P(\mathrm {H0}|x)\), can be as high as \(1-\alpha \).
This means that a significance test can reject a point null hypothesis with a very small p value, although at the same time, there is a very high probability that the null hypothesis is true. The lesson here is that the single p value for the null hypothesis, even when it is extremely small, can be more difficult to interpret than is commonly assumed.
5 Arguments against omnibus tests for comparing classifiers
When the performance of multiple classifiers is compared on more than one data set, it is now common practice to account for multiplicity effects by means of an omnibus test. Here, the global null hypothesis is that there is no difference between any of the classifiers. If the omnibus test gives a significant result, then we may conclude that there is a significant difference between at least one pair of classifiers. A post-hoc test can then be applied to detect which pair(s) are significantly different. The Friedman test is a non-parametric omnibus test for analyzing randomized complete block designs (Friedman 1937, 1940). This test is now widely used for the comparison of multiple classifiers (Demšar 2006), together with the Nemenyi post-hoc test (Nemenyi 1963). However, there are several problems with this approach.
First, in contrast to common belief, the Friedman test is not a non-parametric equivalent of the repeated-measures ANOVA, but it is a generalization of the sign test (Zimmerman and Zumbo 1993; Baguley 2012). This is because the ranks in the Friedman test depend only on the order of the scores (here, observed performance) within each subject (here, data set), but the test ignores the differences between subjects. As a sign test, the Friedman test has relatively low power (Zimmerman and Zumbo 1993). Baguley (2012) advises us that rank transformation followed by ANOVA is both a more powerful and robust alternative.
Second, the Friedman test sacrifices information by requiring that real values are rank-transformed. Sheskin (2007) explains that this is one reason why statisticians are reluctant to prefer this test over parametric alternatives, even if one or more of their assumptions are violated. Furthermore, note that the transformation into ranks depends on the rounding of the real values. For example, assume that three classifiers achieve the following accuracies on one of the data sets: 0.809, 0.803, and 0.801. The corresponding ranks are then 1, 2, and 3. If we round the values to two decimal places, then the ranks are 1, 2.5, and 2.5. It is possible that such ties can change the result from non-significant to significant. It is somehow disconcerting that mere rounding can have such an effect on the outcome of the test.
Third, it is widely assumed that post-hoc tests for multiple comparisons may be conducted only if an omnibus test has first given a significant result. The rationale is that we need to control the family-wise Type I error rate. But there is an alternative view among statisticians that adjustments for multiple testing are not necessarily needed (Rothman 1990; Poole 1991; Savitz and Olshan 1998), and that such adjustments can even create more problems that they solve (Perneger 1998). If the omnibus test is a one-way ANOVA, then post-hoc tests are valid,Footnote 3 irrespective of the outcome of the omnibus test (Sheskin 2007). Hsu (1996) deplores that it has become an unfortunate common practice to pursue multiple comparisons only when the global null hypothesis has been rejected.
Implicit in the application of the Friedman test is the premise that the global null hypothesis is the most important one: unless we can reject it, we are not allowed to proceed with post-hoc tests. Cohen (1990) argues that we already know that the null hypothesis is false because the difference is never precisely 0; hence, “[...] what’s the big deal about rejecting it?” (Cohen 1990, p. 1000). According to Rothman (1990), there is no empirical basis for a global null hypothesis. Following Rothman’s line of thought, let us suppose that we compare two classifiers, X and Y, on a data set D and observe that X is significantly better. Would we then not recommend X over Y for data sets that are similar to D? But now suppose that we apply three classifiers, X, Y, and Z to the data set D. We use an omnibus test (or otherwise correct for multiple testing), and we now fail to reject the global null hypothesis of equal performance. Would we still recommend X over Y despite the lack of significance? The difference in accuracy (or whichever metric we are using) between X and Y has not changed—it has of course nothing to do with Z. A defendant of omnibus tests might say that by making more comparisons, we have to pay a “penalty for peeking” (Rothman 1990, p. 46), i.e., adopt a stricter criterion for statistical significance. But let us consider the following simplified scenario. Alice designs a study to compare the performance of a support vector machine with random forest on a particular data set. She carries out her experiments in the morning and observes that the support vector machine performs significantly better than random forest. No corrections for multiple testing are needed because there are just two classifiers. Out of curiosity, Alice then applies naive Bayes to the same data in the evening. Clearly, Alice’s new experiment has no effect on her earlier experiments, but should she make multiplicity adjustments? This question does not have an obvious answer because it is not clear where the boundaries of one experiment end and those of another one begin; compare (Rothman 1990; Perneger 1998). Our stance is that adjustments for multiple testing are necessary under some circumstances, for instance, in confirmatory studies where we pre-specify a goal (or prospective endpoints). In exploratory studies, however, we recommend reporting unadjusted p values, while clearly highlighting that they result from an exploratory analysis. Comparative classification studies are generally exploratory, as they normally do not pre-specify any prospective endpoints. Thus, omnibus tests are not needed.
Fourth, to apply the Friedman test, we first need to rank the classifiers from “best” to “worst” for each data set. However, how meaningful is it to give a different rank to classifiers whose performances differ only very slightly? For example, in Guyon et al. (2009), the top 20 models from KDD Cup 2009 were analyzed based on the Friedman test. The model with rank 1 scored \(\hbox {AUC} = 0.9092\) on the upselling test set, while the model with rank 20 scored \(\hbox {AUC} = 0.8995\). Is it meaningful to impose such an artificial hierarchy? We believe that this ranking rather blurs the real picture that all top 20 classifiers virtually performed the same.
Fifth, the Friedman test assumes that the subjects (data sets) have been randomly sampled from one superpopulation. It can be questioned whether in practice, data sets are ever randomly sampled. Surely, purely pragmatic reasons, such as availability, at least influence (if not guide) the choice of data sets. While this violation of a basic assumption is probably widely known, it seems to be tacitly ignored. Do we want to show that there is no difference in performance? Or do we want to show that there is? In Sect. 7.1, we illustrate how we can easily tweak the results by considering different combinations of data sets and classifiers.
Sixth, the experimental results that led to the recommendation of the Friedman test are based on the estimation of replicability as a function of p values (Demšar 2006). However, as we discussed in Sect. 4.4, this approach is questionable.
Finally, our last argument is perhaps the most compelling one. Consider the following simplified example, where \(k = 3\) conditions (\(C_1\), \(C_2\), and \(C_3\)) are applied to \(N = 5\) subjects (Table 1).
The numbers reflect the effect of a condition on a subject. The null hypothesis is stated as \(\text {H0:}~\theta _1 = \theta _2 = \theta _3\), i.e., the median of the population that the numbers 3, 2, 1, 3, 2 represent equals the median of the population that the numbers 4, 5, 6, 4, 5 represent, which also equals the median of the population that the numbers 8, 9, 10, 9, 7 represent. When the null hypothesis is true, the sum of ranks of all three conditions will be equal. The p value of the Friedman test is the probability of observing sums of ranks at least as far apart as the observed ones, under the null hypothesis of no difference.
Let us now consider the following (simplified) drug trial. We administer three drugs, \(C_1\), \(C_2\), and \(C_3\) (one after another, allowing for adequate washout phase, etc.), to \(N=5\) patients and measure how well these drugs improve a certain condition. Here, each patient is a subject, and each drug is a condition (Table 1). From the result of the Friedman test, we can make an inference to the population of patients with similar characteristics, which means patients with the same medical condition, age, sex, etc. For example, we might reject the global null hypothesis that all drugs are equally effective, and a post-hoc test might tell us that drug \(C_3\) is significantly better than \(C_1\) and \(C_2\). Thus, we might conclude that \(C_3\) should be given to the target patients. In this scenario, the Friedman test can be used.
However, if the conditions are classifiers and the subjects are benchmark data sets, then we have a problem. Commonly used data sets (e.g., the Transfusion and the King-rook-vs-king-pawn data sets from the UCI repository) are completely diverse entities that cannot be thought of as originating from one superpopulation. There is simply no “population of data sets.” This means that the numbers 3, 2, 1, 3, 2 for condition \(C_1\), for example, cannot be a sample from one population. Unless the subjects originate from the same population, all inferences based on the Friedman test are elusive. In fact, this last argument applies to any omnibus test, not just the Friedman test. Therefore, such tests are inappropriate when the subjects represent diverse data sets.
6 Confidence curve
We now present an alternative method, the confidence curve, which is a variant of Birnbaum’s confidence curve estimator (Birnbaum 1961). Confidence curves enable us to assess simultaneously how compatible an infinite number of null hypotheses are with our experimental results. Thereby, our focus is no longer on “the” null hypothesis of no difference, its single p value, and the question whether it is significant or not. This shift of focus can help us avoid the major problems of NHST and the p value.
6.1 Illustration of key concepts
A confidence curve is a two-dimensional plot that shows the nested confidence intervals of all levels for a point estimate. We consider the effect size, which we define as follows.
Definition 1
(Effect size) Let \(\tau _A\) and \(\tau _B\) be the true performance and \(o_A\) and \(o_B\) be the observed performance of two classifiers, A and B, on data D. The true (unstandardized) effect size is the difference \(\delta = \tau _A - \tau _B\). The observed difference \(d = o_A - o_B\) is the point estimate of \(\delta \).
For example, if a classifier A achieves 0.85 accuracy on a specific data set while a classifier B achieves only 0.70, then the estimated effect size is \(d = 0.15\). Of course, the effect size could be measured based on any performance metric.
Key elements of the confidence curve for the difference in performance between two models. By mentally sliding the red vertical line (“null line”) along the x-axis, we can assess how compatible the corresponding null hypothesis is with the observed data. This compatibility is maximal (\(p\, \mathrm{value} =1.0\)) when the red line reaches d, which corresponds to the null hypothesis H0: \(\delta =d\). As the red line moves away from d in either direction, the corresponding null hypotheses become less and less compatible with the observed data
Figure 3 illustrates the key features of the confidence curve. In this example, the point estimate of the effect size is 0.15. The plot has two complementary y-axes. The left y-axis shows the p value associated with each null value, which is shown on the x-axis. The right y-axis shows the confidence level. In Fig. 3, the p value of “the” null hypothesis of no difference, \(\text {H0:}~\delta = 0\), is 0.15. Each horizontal slice through the curve gives one confidence interval. For example, the 95%-confidence interval for \(\delta \) is the slice through the p value of 0.05. Technically, the maximum of the curve gives the zero percent confidence interval. In this example, the confidence intervals are symmetric; thus, we obtain a “tipi”-shaped symmetric confidence curve around the point estimate.
In Fig. 3, we see that the null value of no difference lies within the 95%-confidence interval. By conventional criteria, we would therefore fail to reject the null hypothesis of no difference. However, note that a confidence curve should not be used as a surrogate significance test. A confidence curve can tell us much more. First and foremost, it disentangles the effect size from the precision of its measurement. The effect size is the magnitude of the observed difference, d, while its precision is given by the width of the curve. The wider the curve, the less precise is the measurement, and the narrower the curve, the more precise is the measurement. The area under the confidence curve (AUCC) can therefore be considered a measure of precision.
Note that every p value is associated with exactly two different null values because we are considering precise point null hypotheses (e.g., \(\text {H0:}~\delta = 0\)). Consider the dotted horizontal line at the p value of 0.15 (Fig. 3). This line crosses the curve at the “entangled” null value (marked by a star), which corresponds to the null hypothesis \(\text {H0:}~\delta = 0.30\) in this example. This means that both the null hypothesis of no difference and the entangled null hypothesis are associated with exactly the same p value. Therefore, there is no reason why we should prefer one null hypothesis over the other; both hypotheses are equally compatible with the data.
Figure 3 shows the infinite spectrum of null hypotheses on the x-axis and how compatible they are with our data at any given level of confidence. Not surprisingly, the null hypothesis \(\text {H0:}~\delta = d\) is most compatible. The left y-axis shows all possible p values. It might perhaps seem surprising to see p values appear in the proposed alternative method, given all the arguments against the p value in Sect. 4. However, note that these arguments pertain to the single p value from one single null hypothesis test. Poole (2001) refers to this p value more precisely as the “null p value” (p. 292). But in contrast to NHST, confidence curves do not give undue emphasis to a single p value.
To check the compatibility of other null hypotheses with the obtained data, one can easily imagine sliding the red “null line” in Fig. 3 along the x-axis and see where it intersects with the confidence curve. Thus, confidence curves allow us to check the compatibility of an infinite number of null hypotheses with our experimental results. Compatibility is a gradual, not a dichotomous characteristic; some hypotheses are more, others are less compatible.
6.2 Confidence curves for the effect size in repeated cross-validation
We consider only one data resampling scheme, r-times repeated k-fold cross-validation because it is probably the most widely used strategy. A \((1-\alpha )100\%\) confidence interval for the true effect size can be derived from the variance-corrected resampled t test (Nadeau and Bengio 2003; Bouckaert and Frank 2004),
where t is the critical value of Student’s distribution with \(\nu =kr-1\) degrees of freedom; k is the number of cross-validation folds; r is the number of repetitions; \(n_2\) is the number of cases in one validation set; and \(n_1\) is the number of cases in the corresponding training set (where \(n_1 \approx 5n_2\)). The standard deviation s is calculated as
where \(d_{ij}\) is the difference between the classifiers in the ith repetition of the jth cross-validation fold, and \(\bar{d}\) is the average of these differences. Note that in the case of repeated cross-validation, the sampling intention is clear. The confidence intervals, and thereby the confidence curve, assume that exactly kr samples were to be taken.
The confidence curve consists of an infinite number of nested confidence intervals for the true effect size. This nesting can be described as follows. Let \(F_{d,\sigma }(x)\) be a cumulative distribution function with density \(f_{d,\sigma }(x)\). The confidence curve c(x, d) is then defined as shown in Eq. (1).
When the degrees of freedom are sufficiently large (\(\nu > 30\)), the t-distribution approximates the standard normal distribution, and F can be approximated by the cumulative distribution function of the normal distribution, \(\Phi \). The difference between Eq. (1) and Birnbaum’s estimator is the factor 2, which is needed for two-sided p values.
The basic algorithm for plotting confidence curves consists of four simple steps, (1) calculating a few dozen confidence intervals at different levels, from 99 to 0%; (2) plotting \(\alpha \) as a function of the lower bound; (3) plotting \(\alpha \) as a function of the upper bound; and (4) interpolating through all points. The following pseudocode plots the confidence curve for a difference in performance that is measured in r times repeated k-fold cross-validation. The supplementary material at https://github.com/dberrar/ConfidenceCurve contains the R code PlotConfidenceCurve.

6.3 Area under the confidence curve
When we compare the performance of many classifiers and there are space limitations, it can be preferable to tabulate the results instead of plotting all confidence curves. Confidence curves can be summarized by two values, the point estimate and their width. The wider the confidence curve, the less precise is the measured performance difference. Thus, the area under the confidence curve (AUCC) can be used as a measure of precision. But clearly, by using a single scalar, we lose important information about the classification performance. For r-times repeated k-fold cross-validation, the area is given by Eq. (2) (see “Appendix” for details).
6.4 Further notes on confidence curves
In this section, we will illustrate how to use confidence curves.
6.4.1 Statistical significance versus effect size and precision
Statistical significance versus effect size and precision. Confidence curves show nested, non-symmetric Quesenberry and Hurst confidence intervals for the difference in error rates on the test set (Berrar and Lozano 2013). a A narrow curve indicating the absence of any strong effect despite a significant result. The null value \(\delta =0\) (red line) lies outside the 95%-CI. The measurement is very precise. b A wide curve indicating that the data are readily compatible with a moderate to a strong effect despite a non-significant result. The null value \(\delta =0\) (red line) lies inside the 95%-CI. The measurement is not very precise
In the example shown in Fig. 4, we consider the error rate as performance measure. The confidence curve in Fig. 4a is a narrow spike, which indicates a highly precise measurement. The null hypothesis of equal performance can be rejected because the null value, \(\delta = 0\), is outside the 95%-CI of [0.00017, 0.00843] for the point estimate \(d = 0.00430\). Note that the upper bound of the 95%-CI is quite close to the null value. Emphasizing the significance would therefore be misleading in this study. The correct interpretation is that the data are not even compatible with a moderate effect.
Figure 4b shows a wider curve, which indicates that the measurement is less precise. The null value lies within the 95%-CI of \([-0.038, 0.231]\) for the point estimate \(d=0.10\). Based on the conventional criterion, we would not reject the null hypothesis, but the curve in Fig. 4b indicates at least a moderate effect. In fact, null values that are readily compatible with the data span across a relatively wide range. Emphasizing the lack of significance would be misleading in this study. The correct interpretation is that the data are compatible with a moderate to a large effect.
In which situations can we expect such confidence curves? Suppose that we compare the performance of two models, A and B, and A is only marginally better than B. Suppose that this difference is in fact truly negligible for all practical applications. By using a sufficiently large test set, however, we can “make” this difference significant. This is a well-known effect that results from increasing the sample size.Footnote 4 Figure 4a shows the result of such an overpowered study. Here, both the training and the test set contain 10,000 cases; 5000 cases belong to the positive class and 5000 belong to the negative class. Each positive case is described by a 10-dimensional feature vector, with elements randomly sampled from \(\mathcal {N}(0,1)\). The features of the negative cases are randomly sampled from \(\mathcal {N}(1.5,1)\). We trained a classification and regression tree (CART) on the training set and then applied it to the test set. This is model A. Model B is also a CART, but trained on a deliberately corrupted training set: the class labels of 50 randomly selected positive and 50 randomly selected negative cases were swapped. To show that this deliberate corruption has a small but negligible effect, we repeated the experiment 1000 times. We observed that the uncorrupted model performs significantly better (\(p < 0.05\), McNemar’s test) than the corrupted counterpart in 171 experiments. In 139 experiments, however, the corrupted model performed significantly better. In the remaining 690 experiments, there was no significant difference between the two models. The mean and median differences in all 1000 experiments were only 0.0023 and 0, respectively. Figure 4a shows an experiment where the uncorrupted model was slightly better.
In the second experiment (Fig. 4b), the learning set contains only 100 cases, each described by 10 numerical attributes. The first 50 cases belong to the positive class and the remaining 50 cases to the negative class. For the positive cases, the attributes take on values randomly from \(\mathcal {N}(0,1)\). For the negative cases, the attributes take on values randomly from \(\mathcal {N}(0.2,1)\). We trained a random forest classifier and implemented the competing model as a fair coin. We expected that random forest would perform better than the coin, but we obtained the following result: the coin made 55 errors, while random forest made 45 errors. This difference is not significant (\(p = 0.20\), McNemar’s test).
The lesson is that a significant result can be misleading if a strong effect is absent. On the other hand, a non-significant result can be meaningful if there is evidence of a strong effect. Confidence curves keep our focus on what really matters: the effect size and its precision.
6.4.2 Replicability versus reproducibility
It is expedient to distinguish between replicability and reproducibility. According to Drummond (2009), replicability means that the exact experimental protocol can be repeated. By contrast, reproducibility means that the same results can be obtained by other experiments. To replicate a classification study, it would be necessary to make publicly available not only the source code and protocol details, but also all resampled data subsets. Although this is possible, as demonstrated by the OpenML project,Footnote 5 we offer for debate whether replicability is really so desirable. We concur with Drummond (2009), arguing that replicability is an impoverished version of reproducibility.
Comparative classification studies of course do not always involve the same data resampling schemes. Suppose that one study investigated the difference between two algorithms on the basis of five times repeated cross-validation, while another study used tenfold stratified cross-validation. The first study failed to detect a significant difference but the second one did not. How can we reconcile these apparently contradictory results? Replicating both studies will not solve the problem because we would obtain the same results as before.
Consider the following example where we compared the accuracy of random forest and CART on the Ionosphere data set (Fig. 5). We used five different resampling strategies, (a) tenfold cross-validation without stratification; (b) 100 times stratified repeated tenfold cross-validation; (c) tenfold stratified cross-validation; (d) five times repeated twofold cross-validation; and (e) one training set with 70% and one test set with 30% cases (split-sampling).
Let us assume that these resampling strategies represent five studies, published by five different research groups. Furthermore, let us assume that all groups use significance testing. Studies (d) and (e) indicate that there is no difference in performance, in contrast to studies (a)–(c). If all groups published their results, then the literature could be deemed inconclusive regarding the difference between the two algorithms for this particular classification problem. Assuming that there were no errors in the original studies, we would not gain any new insights by replicating them
In contrast, confidence curves can reconcile the apparently contradictory results. The confidence curves in Fig. 5 suggest that the individual studies reproduce each other. The curves in Fig. 5 convey essentially the same message. All studies point to a moderate effect, as can be seen in the average of the confidence curves in Fig. 5f. Random forest outperforms CART, and the true difference in accuracy is about 0.06. Thus, the five studies are in fact confirmatory, not contradictory.
Furthermore, the reliance on statistical tests can lead to a publication bias. We speculate that many researchers feel that a study should not be submitted for publication if the result is not significant, and vice versa. Suppose that only the significant results (a)–(c) were published. These studies would then indicate that the effect (i.e., the difference in performance between random forest and CART on the Ionosphere data set) is \(\frac{1}{3}(0.069+0.060+0.068)=0.066\). This value overestimates the true difference, which, based on all experiments, is only 0.058 (Fig. 5f).
Consider now the following scenario. Assume that the confidence curves from Fig. 5 refer to clinical trials on the effectiveness of a drug. Suppose that this drug really has a small but beneficial effect, as shown in Fig. 5a–c. Let us further assume that a new study (Fig. 5e) is conducted. If this new study focused only on significance, then it would erroneously refute the earlier studies (“no significant effect of the drug was observed”). But the correct interpretation is that this new study confirms the previous ones. Rothman et al. (2008) give two real-world examples of clinical trials where such erroneous conclusions were drawn because of the focus on significance. Confidence curves, on the other hand, would most likely have prevented the investigators from misinterpreting their findings.
6.4.3 Comparison with a null model
Instead of constructing confidence curves for the difference between two real classifiers, A and B, we can construct the curves for A and B with a common baseline model or null model. A natural choice for the null model is the majority voter, which predicts each case as a member of the most frequent class. Another possible choice is the empirical classifier, which uses only the class prior information. If the proportion of class c is p in the training set, and the test set contains n cases, then the empirical classifier will classify np test cases as members of class c; these cases are selected at random. For example, assume that the training set contains only two classes, \(c_1\) and \(c_2\), and the class ratio is \(c_1{:}c_2=30{:}70\) in the learning set. Let the test set contain 200 cases. Then \(200 \times 0.3 = 60\) randomly selected test cases will be predicted as members of class \(c_1\) and \(200 \times 0.7 = 140\) randomly selected test cases will be predicted as members of class \(c_2\). One advantage is that the plot now shows how much a real classifier has learned beyond the information provided by the class distribution in the training set. When we compare n classifiers, we do not need to produce \(\frac{1}{2}n(n-1)\) confidence curves for all pair-wise comparisons but only n curves, i.e., A versus null model, B versus null model, etc. When multiple confidence curves are produced in the same study, it is possible to control the family-wise error rate by adjusting \(\alpha \), which leads to a widening of the curves. However, given the arguments in Sect. 5, we advise against such adjustments in comparative classification studies.
Figure 6a shows the confidence curve for the difference between random forest and CART. Random forest achieved an accuracy of 0.796 whereas CART achieved 0.782. The point estimate of the difference is 0.014, which is quite close to the null value 0. The seemingly good performance of around 80% could invite us to speculate that both random forest and CART have learned something from the features, but this is not the case here. The training set contains 1000 cases, each described by a 10-dimensional feature vector of real values from \(\mathcal {N}(0,1)\). The cases were randomly assigned either a positive or negative class label, with a ratio of 20:80. As the features do not discriminate the classes, no classifier is expected to perform better than the majority voter. Figure 6b shows the confidence curve for the difference between random forest and the majority voter. The majority voter performs slightly better than random forest: the point estimate of the difference in accuracy is \(d = o_{RF} - o_{MV} = -0.004\). Thus, for this data set, there is no reason why we should prefer random forest over the null model.
7 Experiments
We will now use confidence curves and significance testing in a real classification study. Let us imagine two researchers, Alice and Bob, who wish to compare the performance of four classifiers over 14 data sets. To interpret their results, Alice decides to use the Friedman test with Nemenyi post-hoc test, whereas Bob chooses confidence curves. Alice and Bob benchmark the same classifiers on the same data sets based on average accuracy in 10-times repeated tenfold stratified cross-validation (Table 2).
The real-world data sets (#1–#10) are from the UCI machine learning repository (Lichman 2013). The synthetic data sets were generated as follows. Synthetic 1 consists of 100 cases, half of which belong to the positive class and the other half to the negative class. Each case is described by a 10-dimensional numerical feature vector, \(\mathbf x =(x_1,x_2,\ldots ,x_{10})\). The values \(x_i\) of the positive cases \(\mathbf x _+\) are randomly sampled from \(\mathcal {N}(0,1)\). The values of the negative cases \(\mathbf x _-\) are sampled from \(\mathcal {N}(0.5,1)\).
Synthetic 2 consists of 100 cases; 30 cases belong to the positive class, and 70 cases belong to the negative class. Each case is described by a 10-dimensional feature vector. All values \(x_i\) are randomly sampled from \(\mathcal {N}(0,1)\); hence, the features do not discriminate the classes.
Synthetic 3 consists of 100 cases; 20 cases belong to the positive class and 80 cases belong to the negative class. Each case is described by a 10-dimensional feature vector. The ten feature values of the negative and positive cases were randomly sampled from \(\mathcal {N}(0,1)\) and \(\mathcal {N}(0.5,1)\), respectively.
Synthetic 4 consists of 100 cases, half of which belong to the positive class and the other half to the negative class. Each case is described by a 20-dimensional feature vector. For cases of the negative class, the first ten feature values \((x_1,x_2,\ldots ,x_{10})\) were randomly sampled from \(\mathcal {N}(0,1)\). For cases of the positive class, the first ten feature values were randomly sampled from \(\mathcal {N}(0.5,1)\). Irrespective of the class, the next 10 features, \((x_{11},x_{12},\ldots ,x_{20})\), were randomly sampled from a uniform distribution \(\mathcal {U}(-1,1)\).
All experiments in Table 2 were carried out in R (R Development Core Team 2009). NB is a naive Bayes classifier (implementation from R package e1071). RF is a random forest (Breiman 2001), and we used the R implementation randomForest with default settings (Liaw and Wiener 2002). CART is a classification and regression tree (Breiman et al. 1984); we used the R implementation rpart (Therneau et al. 2014). EC is an empirical classifier, which uses only the class prior information to make predictions and ignores any covariate information.
7.1 Analysis with Friedman test and Nemenyi posthoc test
The Friedman test statistic is defined as
where N is the number of data sets and k is the number of classifiers; \(R_{j}^2=\frac{1}{N}\sum _{i}r_{i}^j\), where \(r_{i}^{j}\) is the rank of the jth algorithm on the ith data set (Demšar 2006). Iman and Davenport (1980) proposed a less conservative statistic,
which is distributed according to the F-distribution with \(\nu _1 = (k-1)\) and \(\nu _2 = (k-1)(N-1)\) degrees of freedom. For the experimental data (Table 2), Alice obtains
and
For \(\nu _1=4-1\) and \(\nu _2=(4-1)(14-1)\), Alice obtains the critical value of the F-distribution for \(\alpha =0.05\) as \(F(3,39)=2.85\). As \(F_F > 2.85\), Alice concludes that there is a significant difference between the classifiers. She therefore proceeds with Nemenyi post-hoc test to find out between which pairs of classifiers a significant difference exists. Two classifiers perform significantly differently if their average ranks differ by at least the critical value \(CD = |q_\alpha \sqrt{\frac{k(k+1)}{6N}}| = 2.569 \sqrt{\frac{20}{84}} = 1.25\). Alice concludes that all pair-wise comparisons involving EC are significantly different.
Alice wonders whether the difference between any pair of NB, RF, and CART becomes significant without the null model, EC. By discarding EC, Alice obtains \(F_F = 4.48\), which exceeds the critical value of \(F(2,26)=3.369\). Alice therefore applies again Nemenyi post-hoc test and finds that the absolute difference between the average ranks of RF and CART, 0.964, exceeds the new critical difference of 0.886. Alice thinks: “Surely, it cannot be wrong to report this result, as excluding EC has no effect on the ranks of the other classifiers. The reason why I get a significant result now is that the critical difference has decreased.”
Alice wonders what would happen if she focused only on the real-world data sets and the real classifiers. Alice obtains \(F_F = 6.19\), which exceeds the critical value of \(F(2,18) = 3.55\); therefore, Alice proceeds with the post-hoc test. The new critical difference is 1.05. This time, however, the significant difference between RF and CART vanishes because the absolute difference between their average ranks is only 0.85. By contrast, Alice observes now a significant difference between naive Bayes and random forest. This result puzzles Alice: “What would it actually mean for future model selection if I reported that random forest is significantly better than naive Bayes? This result would clearly be misleading: if my new, unseen data sets happen to be similar to the excluded data sets #11–#14, then naive Bayes—and not random forest—seems preferable...”
Alice then considers other combinations of data sets and classifiers. She includes again data sets #11–#14 but excludes data sets #6–#9, compares naive Bayes, random forest, and CART, and obtains \(F_F = 1.50 < 3.55\). The Friedman test now tells her that there is no significant difference. Alice realizes her dilemma: by considering various combinations of data sets and classifiers, she can tweak her results.
What Alice did is not correct, though. As we noted in Sect. 4.1, she should have adhered to everything that she had specified before carrying out her experiments. This means that if she planned to compare four classifiers on 14 data sets, then she cannot change her protocol later; otherwise, the frequentist paradigm is violated and the p value can no longer be interpreted.
This problem is a deep one. In good faith, a researcher might try different combinations of data sets and classifiers. In Guyon et al. (2009), for example, the performance of the classifiers in the KDD Cup 2009 competition was assessed based on the Friedman test. First, the analysis included all submitted classifiers, and then only the (arbitrarily selected?) top 20 classifiers.
All the juggling with data sets and classifiers and the mathematical details above can easily distract us from a more fundamental and far more important question: is an omnibus test actually appropriate here? Unless we are convinced that the data sets in Table 2 originate from one superpopulation, the answer should be “no” (cf. Sect. 5).
7.2 Analysis with confidence curves
Bob decides to compare the performance of the classifiers on each data set individually. For each data set, Bob derives a confidence curve for the performance difference between NB and EC, RF and EC, and CART and EC (Figs. 7, 8, 9). Table 3 shows the corresponding AUCC.
7.2.1 Interpreting confidence curves
Let us consider first the confidence curves for the Sonar data set (Fig. 7a). The cross-validated accuracy is 0.53 for the null model and 0.83 for random forest, so \(d=0.30\). The confidence curve for random forest does not overlap much with the confidence curve of the next best model, CART (blue). However, there is quite some overlap between the curves for CART and naive Bayes (red). Unsurprisingly, all classifiers performed better than the empirical classifier; random forest, however, is the preferable model for this data set, as its confidence curve (green) is most to the right. Note that the null value \(\delta =0\) (vertical line) lies well outside the 95%-CI, which means that random forest performs significantly better than the null model.
On the data sets Spect, Heart, and Pima (Fig. 7b, c, f), we see a large overlap of the confidence curves, indicating that the differences in performance are probably negligible for all practical purposes. Compared to the curves for the Sonar data set, the curves are much narrower, which reflects a higher precision. Bob concludes that all classifiers are suitable for these data sets. Given the large overlap of the curves, Bob also concludes that the performance differences are negligible.
On the data set Ionosphere (Fig. 7d), the confidence curves also show some overlap, but Bob considers them sufficiently far apart to argue that the performance differences matter. His conclusion is that random forest is preferable to CART, which is in turn preferable to naive Bayes.
On the data set Transfusion (Fig. 7e), naive Bayes and random forest performed essentially the same, whereas CART performed remarkably better. Bob cannot explain why CART, a single tree, could outperform an ensemble of trees in this classification task.
In Fig. 8a, the confidence curves paint a clear picture: random forest is preferable to CART, which in turn performs substantially better than naive Bayes on the data set Tic-tac-toe. By contrast, the performance differences are less pronounced in the data set German (Fig. 8b).
For the data set Liver (Fig. 8c), random forest seems preferable to CART: the point estimate for the difference between random forest and the null model is 0.23, while it is 0.17 for the difference between CART and the null model. Bob could carry out a statistical test to assess the difference between random forest and CART. Without adjustment for multiple testing, the variance-corrected resampled t test gives a significant result (\(p \ \text {value} = 0.02\)), but this result adds little to the interpretation. The vertical line at \(\delta =0\) crosses the confidence curve of naive Bayes at the p value of 0.26; hence, naive Bayes does not perform significantly better than the null model. However, at the 95%-confidence level, compatible values for the true difference range from about \(-0.03\) to 0.13, indicating that naive Bayes is preferable to the null model despite the lack of significance.
On the data set KRvsKP (Fig. 8d), random forest is again the preferable model, although its performance is not substantially different from that of CART. Note the narrowness of the confidence curves, which reflects a high precision. With \(N = 3196\), the number of cases is the largest in KRvsKP;Footnote 6 therefore, the performance could be measured most precisely for this data set.
For Synthetic 1 (Fig. 9a), CART achieved a performance of 0.550, which is only marginally better than that of the null model with 0.508. The null value lies well within the 95%-CI, suggesting that the difference is not significant, but we remember that a confidence curve should not be used as a surrogate significance test. Instead, we should consider their shape and position. Note that the curve for CART is relatively wide. Actually, plausible values for the difference in performance range from \(-0.11\) to 0.19 (the bounds of the 95%-CI), which points to at least a moderate effect. Synthetic 1 was created by random sampling from a known distribution. Is it possible that this particular instantiation of the data set was just an “unlucky” sample for CART, and that CART could perform much better than the null model on other instantiations? As we will show later, this is indeed the case. For Synthetic 1 (Fig. 9a), the curve for naive Bayes partly overlaps with that of random forest, but Bob considers the point estimates sufficiently far apart and therefore concludes that naive Bayes is preferable.Footnote 7
The data set Synthetic 2 (Fig. 9b) is random, with a class ratio of 30:70. The expected accuracy of the empirical classifier (null model) is \(0.3 \times 0.3 + 0.7 \times 0.7 = 0.58\). In the experiment, the empirical classifier achieved 0.59. The best a classifier can do is predict like the majority voter, with an expected accuracy of 0.70. Random forest comes closest to this performance with an accuracy of 0.682. The null value lies within the 95%-CI of all models; thus, by conventional reasoning, none of the models would be considered significantly better than the null model. But the width and the position of the curves for naive Bayes and random forest suggest that these models are, in fact, preferable to the null model. By contrast, the difference between CART and the null model is negligible.
On Synthetic 3 (Fig. 9c), naive Bayes performed slightly better than random forest, but given the large overlap of the curves, the difference can be neglected. Although CART does not perform significantly better than the null model, the position and width of its confidence curve suggest that CART is preferable.
On Synthetic 4 (Fig. 9d), naive Bayes performs again better than random forest. Note that the curves are wider than those in Fig. 9c. The reason is that Synthetic 4 contains ten additional, irrelevant features, which contribute to the larger variance in accuracy. None of the models performs better than the null model at the conventional significance level. However, plausible null hypotheses include relatively large values (up to 0.29 for naive Bayes), suggesting a reasonably large effect. Emphasizing the lack of significance would not do justice to the actually good performance.
To further illustrate pair-wise performance differences, let us consider the confidence curves for the difference between naive Bayes and random forest on Synthetic 1 and Synthetic 4. In Fig. 10a, the null value \(\delta =0\) is just outside the 95%-CI, suggesting that naive Bayes performs significantly better than random forest on Synthetic 1. In contrast, the null value is well within the 95%-CI in Fig. 10b. The point estimate of the difference is 0.05. Plausible values for the difference range from \(-0.06\) to 0.16 (the bounds of the 95%-CI). The shape and position of the curve points to a moderately large effect. Both curves in Fig. 10 suggest that naive Bayes is preferable to random forest for these two data sets; significance, or lack thereof, is irrelevant for this conclusion.
Next, we verified whether Bob’s conclusions based on the confidence curves can be justified empirically. As we know the generative functions of the synthetic data sets, we created 1000 new instantiations for each data set and applied the classifiers again. Then, we created box-and-whiskers plots for the resulting cross-validated accuracies (Fig. 11).
Distribution of average accuracy in 10 times repeated tenfold stratified cross-validation for the synthetic data sets. For each data set, 1000 instantiations were created by random sampling from its underlying distribution. Each instantiation was then classified by the four models. Shown are the box-and-whiskers plots of the resulting accuracies of each model. a Synthetic 1: class ratio 50:50, 10 features, features of class 0 from \(\mathcal {N}(0,1)\), features of class 1 from \(\mathcal {N}(0.5,1)\); b Synthetic 2: class ratio 70:30, 10 features, all from \(\mathcal {N}(0,1)\); c Synthetic 3: class ratio 80:20, 10 features, features of class 0 from \(\mathcal {N}(0,1)\), features of class 1 from \(\mathcal {N}(0.5,1)\); d Synthetic 4: class ratio 50:50, 20 features, features 1..10 of class 0 from \(\mathcal {N}(0,1)\), features 1..10 of class 1 from \(\mathcal {N}(0.5,1)\), features 11..20 from \(\mathcal {U}(-1,1)\) irrespective of class
Figure 11a reveals that naive Bayes is indeed preferable to random forest for Synthetic 1. Particularly, the difference between CART and the null model (EC) is striking. CART performs indeed better than the null model. Thus, Bob’s conclusions based on the confidence curve (Fig. 9a) are confirmed.
For data set Synthetic 2, Bob concluded that the difference between CART and the null model was negligible. In contrast, he concluded that both random forest and naive Bayes are suitable for this data set despite their lack of significance. Bob concluded that random forest is slightly better than naive Bayes. Figure 11b confirms Bob’s interpretation: random forest performs indeed slightly better than naive Bayes; however, the interquartile range for random forest lies almost entirely in that for naive Bayes, suggesting that the difference is only marginal. Based on a visual inspection of the box-and-whiskers plots, CART cannot be considered superior to the null model.
Based on the confidence curves in Fig. 9c, Bob concluded that the difference between naive Bayes and random forest is negligible; the box-and-whiskers plots in Fig. 11c support this conclusion. Interestingly, CART performs better than the null model, which confirms Bob’s interpretation based on Fig. 9c. If Bob had based his interpretation on the test, then he would have concluded that CART does not perform significantly better than the null model.
The confidence curves for random forest and CART in Fig. 9d suggest no substantial difference between these two models. However, Fig. 11d suggests that random forest is preferable to CART because the interquartile ranges do not overlap. Here, Bob’s interpretation of the curves conflicts with the interpretation of the box-and-whiskers plots. Figure 11d shows that naive Bayes performs slightly better than random forest on Synthetic 4. Both models perform better than CART, and each model performs substantially better than the null model. This confirms Bob’s interpretation of Fig. 9d. If he had based his verdict on significance only, then he would have considered no model superior to the null model, as the null value lies in all 95%-confidence intervals.Footnote 8
7.2.2 Interpreting the area under the confidence curve
If a study includes many classifiers and data sets, then it may not be feasible to plot all confidence curves because of space limitations. In that case, it can be preferable to tabulate the point estimates of the performance differences with their AUCC (Table 3).
In Table 3, the difference between the null model and naive Bayes is 0.15 for Sonar. We can interpret this value as follows: by using naive Bayes instead of the null model, we gain an additional 15% in accuracy. The area under the confidence curve (AUCC) can be interpreted in terms of the precision of this estimate. In other words, the AUCC is a scalar that represents a numerical estimate of the precision. The closer the value is to 0, the narrower is the confidence curve, and the more precise is the estimate; by contrast, the larger the AUCC, the wider the curve, and the less precise is the estimate.
Let us consider the performance of naive Bayes on Spect and Heart. By using naive Bayes instead of the null model, we gain 12% in accuracy on Spect but nearly three times as much (34%) on Heart. Both estimates have comparable precisions (\(\hbox {AUCC} = 0.053\) and \(\hbox {AUCC} = 0.054\), respectively). We can therefore conclude that naive Bayes is much more suitable to data sets that are similar to the Heart data set. Among all investigated data sets, naive Bayes is most suitable to the data set KRvsKP. The gain in accuracy is 38%, which is also the most precise measurement with \(\hbox {AUCC} = 0.017\).
Similarly, point estimates and AUCC can be compared across classifiers. For example, consider the performance on Tic-tac-toe. Compared to the null model, naive Bayes yields a gain in accuracy of 15% (\(\hbox {AUCC} = 0.033\)), whereas the gain is 36% for CART (\(\hbox {AUCC} = 0.033\)). As the precision is the same, we can be confident that CART is more than twice as suitable to this data set. However, random forest is even better, with a gain in accuracy of 44% (\(\hbox {AUCC} = 0.027\)). The performance of random forest is almost three times better than that of naive Bayes on the data set Tic-tac-toe. All other comparisons can be made analogously.
8 Discussion
We investigated several problems of null hypothesis significance testing for the comparison of classifiers. We discussed that the Fisherian and Neyman–Pearsonian schools of thought are widely believed to originate from one single, coherent theory of statistical inference, although their philosophies are fundamentally different (Goodman 1993; Hubbard and Bayarri 2003; Hubbard and Armstrong 2006). We paid particular attention to the problems of the p value, which have received scant attention in the machine learning literature so far. First, perhaps the most persistent misconception is that the p value is a completely objective measure of statistical inference. Second, the p value is not predictive of reproducibility. Third, the p value confounds the effect size (e.g., the magnitude of the performance difference) and the precision with which that effect has been measured. And finally, the p value invites a dichotomization into significant and non-significant findings and thereby emphasizes decision making over estimation. We argue that estimation, not decision making, should be the proper role for statistical inference in machine learning.
There is certainly no shortage of papers criticizing NHST, notably in psychology and sociology (Rozeboom 1960; Carver 1978; Cohen 1994; Schmidt 1996). On the other hand, it is extraordinarily difficult to find explicit defenses of this practice. Four defenses are particularly noteworthy, though. Levin (1998) argues in favor of significance testing because it can be done intelligently and because alternatives, such as confidence intervals, also have their caveats and pitfalls. In a similar line of defense, Abelson (1997) argues that significance testing can be justified as long as we are only interested in the direction of the finding and not in the magnitude of the effect. Krueger (2001) defends the pragmatic value of NHST while clearly acknowledging its logical deficiencies. Note, however, that these are defenses of (Fisherian) significance testing, not arguments in favor of the Neyman–Pearsonian hypothesis testing or, worse, the hybrid approach. We acknowledge that there is a time and place for significance testing, for example, in feature selection tasks. For the comparison of classifiers, however, our stance is that no such tests are needed.
For several decades, significance testing has fueled heated debates, yet despite these criticisms, it seems to be a widely held view that statistical tests are essential for the interpretation of empirical results in machine learning. Why is that so? We speculate that there are two main reasons. First, many researchers may feel a need for a clear-cut decision (Schmidt and Hunter 1997), which the dichotomous verdict of a test can provide (“significant” versus “non-significant”). Second, well-meaning researchers might believe that scientific integrity necessitates an objective, rigorous procedure, which, supposedly, can be provided by NHST. Statistical tests are indeed often believed to give “certain reassurance about the validity and non-randomness of the published results” (Demšar 2006, p. 27). Apparently, the p value—provided that it is sufficiently small—can give such a reassurance. In this study, we challenge these views. We agree with Berger and Berry (1988) who caution against the illusionary objectivity of statistical testing.
We then discussed the problem of multiple comparisons in classification studies. It may be widely assumed that multiple comparisons generally require that p values be adjusted; however, there is no consensus among statisticians that this is indeed so (Poole 1991; Rothman 1990). Researchers who argue in favor of adjustments are faced with the difficult choice from a wealth of procedures, ranging from the very conservative Bonferroni correction to more advanced methods; see, for example, (García and Herrera 2008). We provided several arguments against omnibus tests in comparative classification studies; in particular, we questioned the appropriateness of the Friedman test for this task.
As an alternative to NHST, we propose confidence curves. A key problem of classic statistical testing is the focus on one single null hypothesis, i.e., the null hypothesis of no difference, and its p value. In contrast, confidence curves do not put undue emphasis on that hypothesis as the only one of interest. Instead, confidence curves enable us to easily check how compatible other hypotheses are with our experimental results. In fact, we can mentally check the compatibility of an infinite number of null hypotheses. Compatibility should not be regarded as a dichotomous but as a gradual characteristic; some hypotheses are more, others are less compatible. Whether a difference is significant or not can be easily verified by checking where the null line intersects the confidence curve, but doing so would defeat all the advantages of confidence curves over significance tests. Significance (or lack thereof) should not be our major concern. A confidence curve brings to the forefront what matters: the effect size and its precision. We think that this is in the spirit of F. Yates when he wrote that
[...] scientific research workers [...] pay undue attention to the results of the tests of significance [...] and too little to the estimates of the magnitude of the effects they are investigating. (Yates 1951, p. 32)
Our study has several limitations, though. First, we considered only one performance metric, accuracy, and only one data resampling strategy (repeated cross-validation). Our future work will focus on other performance measures and data resampling strategies.
Second, the use and comparison of confidence curves might be criticized as lacking objectivity. However, a confidence curve is merely a tool that summarizes the results, putting emphasis on the effect size and its reasonable bounds, and it is incumbent on the investigator to construct an argument for or against a classifier based on that curve. The investigator’s informed judgment is a crucial element in the interpretation of data (Cohen 1994). Clearly, this judgment may not always be convincing to everyone. For example, a researcher might consider the point estimate of the difference in performance sufficiently large and the curve sufficiently narrow to conclude that one model is superior to another one for a concrete classification task. Not all readers may agree with this particular judgment, and that is fine. They can judge by themselves whether the investigator’s reasoning is plausible or not and then draw their own conclusions. By contrast, how would a researcher make her case with a NHST? Her argument would stand or fall with a single p value, requiring no further intellectual engagement, which Cox and Hinkley (1974) criticized as follows:
It is essentially consideration of intellectual economy that makes a pure significance test of interest. (Cox and Hinkley 1974, p. 81)
When Bob refrains from making a decision regarding performance differences, Alice might object, demanding a final, clear-cut verdict. But are categorical statements always possible—or even desirable—in the scientific analysis of data? Poole (1987) reminded us that science and decision making are two different enterprises: sciences focuses on learning, explaining, and understanding, whereas decision making focuses on reasons to act or refrain from acting. In a similar vein, Rothman et al. (2008) consider estimation, and not decision making, as the proper role for statistical inference in science. While there may not be a consensus among statisticians about the proper role of statistical inference in data analysis, we posit that machine learning would benefit from a stronger focus on estimation and not decision making. This does of course not imply that test statistics have no role to play in machine learning; for example, for feature selection from high-dimensional data sets, appropriate test statistics can of course be extremely useful.
Finally, like confidence intervals, confidence curves need to be interpreted within the frequentist framework. There exist alternative methods, such as likelihood ratios (Cox 1958; Goodman and Royall 1988), which directly measure the weight of evidence for and against hypotheses. But like confidence curves, these alternative methods are no silver bullet. We doubt that there is one. Evaluation tools should assist the investigator in interpreting the experimental results and support, not supplant, informed judgment. We believe that confidence curves, despite their limitations, serve this purpose well.
9 Conclusions
Null hypothesis significance testing has become deeply entrenched in machine learning for the comparison of classifiers. A significance test is widely considered necessary to underpin the sound and objective interpretation of empirical results. However, this practice provides only a veneer of rigor. A thorough interpretation of classification results does not need to rely on significance tests. We conclude that null hypothesis significance testing should be replaced by methods that support informed judgment and put greater emphasis on effect size estimation, and not on decision making. The use of the proposed confidence curves could be a step in this direction. We hope that our study will encourage a widespread debate on the role of statistical tests in machine learning and spur further research on alternative evaluation methods and visualization tools that give more room to informed judgment.
Notes
The standard t test is not applicable because the individual training sets overlap, so the independence assumption is violated, which leads to an underestimation of the true variance. This is what the term \(\frac{n_2}{n_1}\) corrects.
The two exceptions are Scheffe’s test and Fisher’s Least Significant Difference (LSD) test.
“Virtually any study can be made to show significant results if one uses enough subjects regardless of how nonsensical the content may be.” (Hays 1963, p. 326).
For Tic-tac-toe, \(N = 958\); for German, \(N = 1000\); for Liver, \(N = 345\).
As we will show in Fig. 10a, this difference is also significant.
And if Bob had corrected for multiple testing, then the results would have been even “less significant”.
References
Abelson, R. (1997). A retrospective on the significance test ban of 1999 (if there were no significance tests, they would need to be invented). In L. Harlow, S. Mulaik, & J. Steiger (Eds.), What if there were no significance tests? (pp. 117–141). Mahwah, NJ: Psychology Press.
Baguley, T. (2012). Serious stats: A guide to advanced statistics for the behavioral sciences. New York: Palgrave Macmillan.
Bakan, D. (1966). The test of significance in psychological research. Psychological Bulletin, 66(6), 423–437.
Bayarri, M., & Berger, J. (2000). P values for composite null models. Journal of the American Statistical Association, 95(452), 1127–1142.
Benavoli, A., Corani, G., Mangili, F., & Zaffalon, M. (2015). A Bayesian nonparametric procedure for comparing algorithms. In Proceedings of the 32nd international conference on machine learning, JMLR.org, JMLR Proceedings (Vol. 37, pp. 1264–1272).
Berger, J., & Berry, D. (1988). Statistical analysis and the illusion of objectivity. American Scientist, 76(2), 159–165.
Berger, J., & Delampaday, M. (1987). Testing precise hypotheses. Statistical Science, 2(3), 317–352.
Berger, J., & Sellke, T. (1987). Testing a point null hypothesis: The irreconcilability of \(p\) values and evidence. Journal of the American Statistical Association, 82, 112–122.
Berrar, D., & Lozano, J. (2013). Significance tests or confidence intervals: Which are preferable for the comparison of classifiers? Journal of Experimental and Theoretical Artificial Intelligence, 25(2), 189–206.
Berry, D. (2006). Bayesian clinical trials. Nature Reviews Drug Discovery, 5, 27–36.
Birnbaum, A. (1961). A unified theory of estimation. I. Annals of Mathematical Statistics, 32, 112–135.
Bouckaert, R., & Frank, E. (2004). Evaluating the replicability of significance tests for comparing learning algorithms. In Proceedings of the 8th Asia-Pacific conference on advances in knowledge discovery and data mining, Springer Lecture Notes in Computer Science (Vol. 3056, pp. 3–12).
Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32.
Breiman, L., Friedman, J., Olshen, R., & Stone, C. (1984). Classification and regression trees. New York: Chapman and Hall.
Carver, R. (1978). The case against statistical significance testing. Harvard Educational Review, 48(3), 378–399.
Cohen, J. (1990). Things I have learned (so far). American Psychologist, 45(12), 1304–1312.
Cohen, J. (1994). The earth is round (p \(<\).05). American Psychologist, 49(12), 997–1003.
Corani, G., Benavoli, A., Mangili, F., & Zaffalon, M. (2015). Bayesian hypothesis testing in machine learning. In Proceedings of 2015 ECML-PKDD, Part III, Springer Lecture Notes in Artificial Intelligence (pp. 199–202).
Cox, D. (1958). Some problems connected with statistical inference. Annals of Mathematical Statistics, 29(2), 357–372.
Cox, D. (1977). The role of significance tests. Scandinavian Journal of Statistics, 4(2), 49–70.
Cox, D., & Hinkley, D. (1974). Theoretical statistics. New York: Chapman and Hall/CR.
Cummings, G. (2012). Understanding the new statistics: Effect sizes, confidence intervals, and meta-analysis. New York, London: Routledge, Taylor & Francis Group.
Demšar, J. (2006). Statistical comparisons of classifiers over multiple data sets. Journal of Machine Learning Research, 7, 1–30.
Demšar, J. (2008). On the appropriateness of statistical tests in machine learning. In Proceedings of the 3rd workshop on evaluation methods for machine learning, in conjunction with the 25th international conference on machine learning (pp. 1–4).
Dietterich, T. G. (1998). Approximate statistical tests for comparing supervised classification learning algorithms. Neural Computation, 10, 1895–1923.
Drummond, C. (2006). Machine learning as an experimental science, revisited. In Proceedings of the 21st national conference on artificial intelligence: Workshop on evaluation methods for machine learning, Technical Report WS-06-06 (pp. 1–5). AAAI Press.
Drummond, C. (2009). Replicability is not reproducibility: Nor is it good science. In Proceedings of evaluation methods for machine learning workshop at the 26th international conference on machine learning, Montreal (pp. 1–6).
Drummond, C., & Japkowicz, N. (2010). Warning: Statistical benchmarking is addictive. Kicking the habit in machine learning. Journal of Experimental and Theoretical Artificial Intelligence, 2, 67–80.
Fisher, R. (1943). Note on Dr. Berkson’s criticism of tests of significance. Journal of the American Statistical Association, 38, 103–104.
Fisher, R. (1955). Statistical methods and scientific induction. Journal of the Royal Statistical Society, Series B, 17(1), 69–78.
Folks, J. (1981). Ideas of statistics. New York: Wiley.
Fraley, R., & Marks, M. (2007). The null hypothesis significance testing debate and its implications for personality research. In R. Robins, R. Fraley, & R. Krueger (Eds.), Handbook of research methods in personality psychology (pp. 149–169). New York: Guilford.
Friedman, M. (1937). The use of ranks to avoid the assumption of normality implicit in the analysis of variance. Journal of the American Statistical Association, 32(200), 675–701.
Friedman, M. (1940). A comparison of alternative tests of significance for the problem of \(m\) rankings. Annals of Mathematical Statistics, 11(1), 86–92.
García, S., & Herrera, F. (2008). An extension on statistical comparisons of classifiers over multiple data sets for all pairwise comparisons. Journal of Machine Learning Research, 9, 2677–2694.
Gigerenzer, G. (1998). We need statistical thinking, not statistical rituals. Behavioral and Brain Sciences, 21, 199–200.
Gigerenzer, G., Krauss, S., & Vitouch, O. (2004). The null ritual–What you always wanted to know about significance testing but were afraid to ask. In D. Kaplan (Ed.), The sage handbook of quantitative methodology for the social sciences (pp. 391–408). Thousand Oaks, CA: Sage.
Goodman, S. (1993). \(p\) values, hypothesis tests, and likelihood: Implications for epidemiology of a neglected historical debate. American Journal of Epidemiology, 137(5), 485–496.
Goodman, S. (1999). Toward evidence-based medical statistics. 1: The \(p\) value fallacy. Annals of Internal Medicine, 130(12), 995–1004.
Goodman, S. (2008). A dirty dozen: Twelve \(p\)-value misconceptions. Seminars in Hematology, 45(3), 135–140.
Goodman, S., & Royall, R. (1988). Evidence and scientific research. American Journal of Public Health, 78(12), 1568–1574.
Greenwald, A., Gonzalez, R., Harris, R., & Guthrie, D. (1996). Effect sizes and \(p\) values: What should be reported and what should be replicated? Psychophysiology, 33(2), 175–183.
Guyon, I., Lemaire, V., Boullé, M., Dror, G., & Vogel, D. (2009). Analysis of the KDD Cup 2009: Fast scoring on a large Orange customer database. In JMLR: Workshop and conference proceedings (Vol. 7, pp. 1–22).
Harlow, L., Mulaik, S., & Steiger, J. (1997). What if there were no significance tests? Multivariate applications book series. Mahwah, NJ: Lawrence Erlbaum Associates Publishers.
Hays, W. (1963). Statistics. New York: Holt, Rinehart and Winston.
Hsu, J. (1996). Multiple comparisons: Theory and methods. Boca Raton, FL: CRC Press.
Hubbard, R. (2004). Alphabet soup—blurring the distinctions between \(p\)’s and \(\alpha \)’s in psychological research. Theory and Psychology, 14(3), 295–327.
Hubbard, R., & Armstrong, J. (2006). Why we don’t really know what “statistical significance” means: A major educational failure. Journal of Marketing Education, 28(2), 114–120.
Hubbard, R., & Bayarri, M. (2003). P values are not error probabilities. Technical Report University of Valencia; Accessed 22 Sept. 2016 http://www.uv.es/sestio/TechRep/tr14-03
Hubbard, R., & Lindsay, R. (2008). Why \(p\) values are not a useful measure of evidence in statistical significance testing. Theory Psychology, 18(1), 69–88.
Iman, R., & Davenport, J. (1980). Approximations of the critical region of the Friedman statistic. Communications in Statistics—Theory and Methods, 9(6), 571–595.
Killeen, P. (2004). An alternative to null hypothesis significance tests. Psychological Science, 16(5), 345–353.
Krueger, J. (2001). Null hypothesis significance testing—On the survival of a flawed method. American Psychologist, 56(1), 16–26.
Levin, J. (1998). What if there were no more bickering about statistical significance tests? Research in the Schools, 5(2), 43–53.
Liaw, A., & Wiener, M. (2002). Classification and regression by randomforest. R News, 2(3), 18–22. http://CRAN.R-project.org/doc/Rnews/
Lichman, M, (2013). UCI machine learning repository. University of California, Irvine, School of Information and Computer Sciences. http://archive.ics.uci.edu/ml
Lindley, D. (1957). A statistical paradox. Biometrika, 44, 187–192.
Morgan, P. (2003). Null hypothesis significance testing: Philosophical and practical considerations of a statistical controversy. Exceptionality, 11(4), 209–221.
Nadeau, C., & Bengio, Y. (2003). Inference for the generalization error. Machine Learning, 52, 239–281.
Nemenyi, P. (1963). Distribution-free multiple comparisons. Ph.D. thesis, Princeton University, Princeton.
Neyman, J., & Pearson, E. (1933). On the problem of the most efficient tests of statistical hypotheses. Philosophical Transactions of the Royal Society of London Series A, 231, 289–337.
Nuzzo, R. (2014). Statistical errors. Nature, 506, 150–152.
Perneger, T. (1998). What’s wrong with Bonferroni adjustments. British Medical Journal, 316, 1236–1238.
Poole, C. (1987). Beyond the confidence interval. American Journal of Public Health, 2(77), 195–199.
Poole, C. (1991). Multiple comparisons? No problem!. Epidemiology, 4(2), 241–243.
Poole, C. (2001). Low p-values or narrow confidence intervals: Which are more durable? Epidemiology, 12(3), 291–294.
R Development Core Team. (2009). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org, ISBN 3-900051-07-0
Rothman, K. (1990). No adjustments are needed for multiple comparisons. Epidemiology, 1(1), 43–46.
Rothman, K. (1998). Writing for Epidemiology. Epidemiology, 9(3), 333–337.
Rothman, K., Greenland, S., & Lash, T. (2008). Modern epidemiology (3rd ed.). Philadelphia: Wolters Kluwer.
Rozeboom, W. (1960). The fallacy of the null hypothesis significance test. Psychological Bulletin, 57, 416–428.
Rozeboom, W. (1997). Good science is abductive, not hypothetico-deductive. In L. Harlow, S. Mulaik, & J. Steiger (Eds.), What if there were no significance tests? (pp. 132–149). Mahwah, NJ: Psychology Press.
Savalei, V., & Dunn, E. (2015). Is the call to abandon \(p\)-values the red herring of the replicability crisis? Frontiers in Psychology, 245(6), 1–4.
Savitz, D., & Olshan, A. (1998). Describing data requires no adjustment for multiple comparisons: A reply from Savitz and Olshan. American Journal of Epidemiology, 147(9), 813–814.
Schervish, M. (1996). \(P\) values: What they are and what they are not. The American Statistician, 50(3), 203–206.
Schmidt, F. (1996). Statistical significance testing and cumulative knowledge in psychology: Implications for training of researchers. Psychological Methods, 1(2), 115–129.
Schmidt, F., & Hunter, J. (1997). Eight common but false objections to the discontinuation of significance testing in the analysis of research data. In L. Harlow, S. Mulaik, & J. Steiger (Eds.), What if there were no significance tests? (pp. 37–64). Mahwah, NJ: Psychology Press.
Sellke, T., Bayarri, M., & Berger, J. (2001). Calibration of \(p\) values for testing precise null hypotheses. The American Statistician, 55(1), 62–71.
Sheskin, D. (2007). Handbook of parametric and nonparametric statistical procedures (4th ed.). London/New York: Chapman and Hall.
Stang, A., Poole, C., & Kuss, O. (2010). The ongoing tyranny of statistical significance testing in biomedical research. European Journal of Epidemiology, 25, 225–230.
Sullivan, K., & Foster, D. (1990). Use of the confidence interval function. Epidemiology, 1(1), 39–42.
Therneau, T., Atkinson, B., & Ripley, B. (2014). rpart: Recursive partitioning and regression trees. http://CRAN.R-project.org/package=rpart, R package version 4.1-5.
Thompson, B. (1999). If statistical significance tests are broken/misused, what practices should supplement or replace them? Theory & Psychology, 9(2), 165–181.
Tukey, J. (1991). The philosophy of multiple comparisons. Statistical Science, 6(1), 100–116.
Yates, F. (1951). The influence of statistical methods for research workers on the development of the science of statistics. Journal of the American Statistical Association, 46(253), 19–34.
Zimmerman, D., & Zumbo, B. (1993). Relative power of the Wilcoxon test, the Friedman test, and repeated-measures ANOVA on ranks. The Journal of Experimental Education, 62(1), 75–86.
Acknowledgements
We thank the three anonymous reviewers very much for their detailed and constructive comments that have greatly helped improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editors: Nathalie Japkowicz and Stan Matwin.
Appendix
Appendix
To derive the \(\mathrm {AUCC_{rCV}}\) from r-times repeated k-fold cross-validation, we proceed as follows. We assume that the cumulative distribution function F can be approximated by \(\Phi \). The point estimate d indicates only the location on the x-axis and is irrelevant for the area. Consider Fig. 12. The AUCC for an arbitrary point estimate (\(d = 0.15\) in Fig. 12a) is twice the red area in Fig. 12b. By exploiting the symmetry and choosing \(d = 0\), integration by parts gives
It is obvious that the second term goes to \(\frac{4}{\sqrt{2\pi }}\sigma \) when u goes to infinity. Applying L’Hôpital’s rule, we see that the first term goes to zero when u goes to infinity:
Using the correction term \(\frac{n_2}{n_1}\) for the variance (Nadeau and Bengio 2003; Bouckaert and Frank 2004), we obtain the area under the confidence curve for r-times repeated k-fold cross-validation as
Rights and permissions
About this article
Cite this article
Berrar, D. Confidence curves: an alternative to null hypothesis significance testing for the comparison of classifiers. Mach Learn 106, 911–949 (2017). https://doi.org/10.1007/s10994-016-5612-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10994-016-5612-6