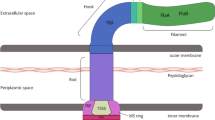
Abstract
Acinetobacter baumannii is an important pathogen responsible for nosocomial infections worldwide. Trimeric autotransporters, the obligate homotrimeric adhesins, are involved in adherence of bacterial cells to various surfaces. These sorts of adhesins were shown to be expressed by A. baumannii. Trimeric autotransporters are modular virulence factors, containing numerous domains and structural architectures. The better understanding of the sequence and structural features of virulence factors are crucial in designing new therapeutic strategies. In this regard, with the aid of reliable in silico tools and the concept of “inherence through homology”, some sequence and structural features of AtaA.baumannii were unveiled. Domain architectures such as the position of repetitive modules, and coiled-coils, along with the prediction of tertiary and quaternary structures, allows us to define some important landscapes of AtaA.baumannii virulence factor. In addition, through CLANS analysis of TAA sequences of Moraxellaceae family, it was concluded that Ata contains several conserved of structural blocks of TAAs. The protein is initiated with extended signal peptide region, a stalk of several head domains and a membrane anchoring region. The globular heads are connected to each other by neck mediators and coiled-coil regions. Several fibronectin and collagen binding sites were defined within the structures. Minimal and maximal frustrated contacts are distributed within the structure of Ata, which suggest both the flexibility and toughness. These are come together for creating an efficient adhesin which is able to bind and bend through multiple sites. The protein could be great target for designing new vaccines or anti-virulence drug.
Similar content being viewed by others
Introduction
Acinetobacter baumannii is a gram-negative, non-fermentative coccobacillus, belonging to Moraxellaceae family. A. baumannii has recently been emerged as a serious health threat, especially in health care units all over the world. The related infections are mostly acquired in hospital settings; however, several reports showed the occurrence of infections in non-hospital environments (Martín-Aspas et al. 2018). Respiratory tract infections, especially in those patients need ventilators, are common. However, the pathogen is able to infect multiple anatomic sites (Harding et al. 2018) such as urinary tract, skin, and soft tissue (Jahangiri et al. 2018; McConnell et al. 2013).
The most important property of A. baumannii is related to its great ability to resist multiple antibiotics (Hsu et al. 2017). The other mentionable property of the bacterium is its fortitude to survive in harsh environments for a long period of time (Rodríguez-Baño et al. 2009).
Despite their high economic burden and the significant reduction in the health-related quality of life of the infected patients, there is no unique virulence factor ascribed to A. baumannii infections (Harding et al. 2018). Expression of complicated regulatory circuitry (Wood et al. 2018), versatile metabolic pathways (Imperi et al. 2011), biofilm formation (De Gregorio et al. 2015) and extraordinary ability to obtain alien genes (Marchaim et al. 2017) are among the mechanisms assisting the organism to persist in harsh environments or initiating the infection.
Among the few virulence factors, trimeric autotransporters (TAAs) are notable which were first introduced in the clinical strains of A. baumannii (Bentancor et al. 2012).
These virulence factors are obligate homotrimers, involving in adhesion of bacterial cells to various surfaces (Cotter et al. 2005); although they are disparate in domains and sequence lengths, the architecture of TAAs generally obeys a simple rule of N-terminal signal peptide, stalk or passenger domain, and a C-terminal membrane anchoring pore (Linke et al. 2006). The term of autotransporter is coined for the sort of such adhesins due to the special property of outer-membrane pore or translocator domain, to secrete the rest of unfolded chains. The process is initiated after the translocation of protein chains from the cytoplasm to the periplasm, where the translocator domain is inserted to the outer membrane with the aid of the β-barrel assembly machinery complex (Schneewind and Missiakas 2014) and projects the passenger to the extracellular environment.
AtaA. baumannii, a TAA of A. baumannii strain ATCC 17978, possesses multiple functions, which are contributed to the multiple domains with intrinsic structures. The adhesin is implemented in adherence of the bacterium to surface components of host epithelial cells like fibronectin and collagen (Weidensdorfer et al. 2016); therefore, it involves in the infection onset and promoting in vivo biofilm formation. The other homolog of TAA has been found in Acinetobacter sp. Tol5 (Koiwai et al. 2016) which has been shown to be essential in high adhesiveness of the bacterium to abiotic surfaces.
Adhesins play central roles in the virulence process, thus identification and characterization of these virulence factors are essential to determine the functional properties of the target protein, to develop preventive and/or therapeutic strategies (Czibener and Ugalde 2012) and provide opportunities for designing new drug targets. In addition, the initial step in understanding the host–pathogen interaction, the objective of molecular infection biology (Diard and Hardt 2017), is determining the structure of virulence factors and their properties. In recent years several members of TAAs have been identified with extensive detail. As crystal structures were solved for many domains of TAAs, it becomes possible to identify the domains of new members by homology modeling approach.
By clustering the sequences of TAAs, related to Moraxellacea family it was shown that AtaA. baumannii comprises several common domains of TAAs. These common domains appear to be conserved in Acinetobacter species, thus we pierce into the structure of these prototypic blocks of AtaA. baumannii inferring from the homology modeling approach (Lee et al. 2007). According to the obtained results, it is suggested that these traits could be extended to most TAAs of the Acinetobacter genus.
Methods
Data Sources
Sequences were obtained from Uniprot knowledge base (Consortium 2017) and Interpro (Finn et al. 2017), the Protein sequence analysis and classification resource. Sequences were collected in CLC sequence viewer 8 (Workbench, Bio 2017) for primary edition and further analyses.
Clustering the TAAs of Moraxellaceae Family
Membrane anchoring region could be used for demarcating the family, thanks to its conservancy across all TAAs (Bassler et al. 2015; Rahbar et al. 2019). Therefore, in the present study, the transmembrane region was served as a query for extracting available sequence data InterPro (Finn et al. 2017) database by querying the YadA like membrane anchoring domain (IPR005594). The extracted sequences, including 551 sequences, were clustered by CLANS software (Frickey and Lupas 2004) employing the all-against-all BLAST approach at https://toolkit.tuebingen.mpg.de (Alva et al. 2016).
Sequence-based Analysis of AtaA. baumannii
The full-length sequence of AtaA. baumannii (under UniProtKB accession number of A3M3H0) was analyzed for determining any repeat module and low complexity regions by Dotlet software at https://dotlet.vital-it.ch. Repetitive modules were also searched within the primary sequence of AtaA.baumannii by the HHrepId (Biegert and Söding 2008) at https://toolkit.tuebingen.mpg.de (Alva et al. 2016). Signal peptide and cleavage site locations were evaluated by SignalP Ver. 4 (Petersen et al. 2011) server at http://www.cbs.dtu.dk. Locations of coiled-coil regions were defined by waggawagga (Simm et al. 2015) at http://waggawagga.motorprotein.de; the server implemented six external tools to extract the coiled-coil regions. All alignments were generated by MUSCLE algorithm (Edgar 2004), implemented in CLC sequence viewer ver 8.
Structure-Based Analyses
Secondary Structure Prediction
Secondary structure prediction of AtaA. baumannii was based on two software tools viz. RaptorX (Wang et al. 2016) at http://raptorx.uchicago.edu and PSIPRED (McGuffin et al. 2000) at http://globin.bio.warwick.ac.uk/psipred/. Additionally, a batch analysis for predicting the secondary structures of all TAAs of Acinetobacters was conducted by RaptorX.
Tertiary Structure Prediction
A multistep approach was employed for building the tertiary structures; briefly, the true available structural templates, homolog to the specific portions of the sequence were selected; the structural homologs were served as templates for building multiple monomers by homology modeling; the best-qualified monomers were selected for further refinement processes; and finally, monomeric structures were used for superimposing to a quaternary structure.
In order to find homologous sequences with known structures, pairwise query-template sequence alignments, based on a comparison of profile hidden Markov models (HMMs) was done by HHpred (Söding et al. 2005) against alignment databases, (these are Pfam and SMART (Letunic and Bork 2017)).
True homologs were defined as structures with probability parameter of 95% or greater (the probability parameter, which provided by the server, should be 95% or greater for a template structure for being homologous to the query sequence).
The selected templates for specific portions of AtaA. baumannii were used for building the related tertiary structures by MODELLER (Webb and Sali 2014). The resulting monomeric structures were refined by ModRefiner (Xu and Zhang 2011) as provided by the Zhang lab at https://zhanglab.ccmb.med.umich.edu.
Refined monomeric structures of AtaA. baumannii were submitted to the GalaxyHomomer (Baek et al. 2017) server at http://galaxy.seoklab.org. This server builds quaternary structures by homology modeling approach. The method was initiated by structure similarity-based approach and ab initio docking to build homomers. The oligomeric state of structures was set as 3. The built trimers underwent additional refinement based on the selected template. The overall quality of all models in all stages was assessed by the MolProbity (Chen et al. 2010) software tool at http://molprobity.biochem.duke.edu. In each step, the best-qualified models were selected for further analyses.
Conservancy Through the Structures
In order to compare sequences for finding conserved residues among TAAs of Moraxellaceae, 555 sequences containing YadA like C-terminal (IPR005594), which were extracted from Interpro database, were aligned by MUSCLE algorithm (Edgar 2004) at the MPI Bioinformatics server (Zimmermann et al. 2017). The alignment file was analyzed and curated by CLC-sequence viewer Ver. 8 (Workbench, Bio 2017). Afterward, the aligned sections corresponding to the predicted structures were used for mapping conservancy on the predicted tertiary structures by Protskin (Ritter et al. 2004).
Membrane Anchoring Region of AtaA. baumannii
Geometrical and physicochemical properties of the membrane anchoring region of the protein were analyzed and visualized by ChexVis (Masood et al. 2015) at http://vgl.serc.iisc.ernet.in using default parameter settings.
For making a comparison, the sequence of membrane anchoring region of AtaA. baumannii was aligned with related regions of ten well annotated TAAs from other genera. Neighbor joining tree on this alignment was also generated by CLC-workbench 8 and visualized by iTOL V4 (Interactive Tree of Life) at https://itol.embl.de.
Energetic State of AtaA. baumannii Structures
Distribution of maximum and minimum frustrations was determined based on the frustratometer ver. 2 tool (Parra et al. 2016) at http://frustratometer.qb.fcen.uba.ar. Local densities of the amino acids were calculated through sequence separation by a distance equal to 12.
B-factors were estimated at https://zhanglab.ccmb.med.umich.edu by ResQ online software (Yang et al. 2016). The method estimated B-factor and residue-level quality in protein structures based on local variations of modeling simulations and the uncertainty of homologous alignments. A set of homologous structures to the query structure were identified by threading and alignment techniques. Then support vector regression was employed for residual error reporting. The B-factor of models was inferred from the experimental records of the top homologous structures.
Results
Sequence Features of Ata
An 1873 amino acid sequence represents a trimeric autotransporter adhesin in Acinetobacter baumannii (strain ATCC 17978/CIP 53.77/LMG 1025/NCDC KC755/5377, Uniprot Acc. No. A3M3H0). In order to understand the sequence features of AtaA. baumannii, properties of signal peptide, location of repeated modules and coiled-coils were determined.
The sequence initiated at N-terminal by an extended signal peptide region (ESPR; Interpro ID: IPR024973). The ESPR is divided into the conserved N-terminal charged domain followed by a conserved hydrophobic domain and a more degenerated region at carboxyl moiety (Fig. 1), containing a cleavage site at Gly54 (Fig. S1). The rest of the sequence included a passenger block of various domains, which terminated in a membrane anchoring region.
Extended signal peptide region (ESPR) properties. Visualization of alignment of 423 ESPR sequences from Pfam with Alvis, sequences are presented as blue bundles, the red line represents the ESPR portion of Ata sequence. All amino acids are sorted based on the guide bar on the left. Polarity and hydrophobicity of sequences are evident in the image
Three compositional biases were observed in the sequence: 385-698 Gly-rich, 557-720 Ala-rich, 775-881 Asn-rich (Fig. 2). Various repeat modules were also observed in several regions of the sequence, mainly the N-terminus region of the protein (Fig. 2). The identified domains at the Pfam database were listed in Table 1.
Dot plot matrix of residues in the Ata whole sequence. The protein sequence (Ata) is compared to itself via DOTLET using the Blosum62 matrix, with a sliding window of 15 amino acids. Symmetric lines forming a square box along the central diagonal indicate the sub-repeats related to two head domains (120–736). The other square (780–1100) indicates the sub-repeats related to GANG and GIN domains. The rest (1103–1773) shows the repeating modules lie on the C-terminal moiety of Ata representing different sizes and numbers of repeats
Several coiled-coil regions were detected within AtaA. baumannii sequence (Table 2), which were different in length and heptad periodicity; well-defined motives of coiled-coils such as IxxNTxx (where x is any amino acid) were also observed. A sample of coiled-coil regions and Asparagine at the fourth position of heptad coiled-coil (N@d) is illustrated in Fig S2.
Structural Features
Secondary Structure Predictions
Secondary structure of the protein was predicted from its amino acid sequence to get insight into the overall structural properties of the protein. The predicted secondary structure components were also employed for controlling the quality of tertiary structures at the next stage. The majority of secondary structure components were predicted to be coil (66%). The extended strand in the beta ladder is the other component of secondary structure for AtaA. baumannii (23%). The other abundant secondary structure for the protein was alpha helixes, which comprised up to 10% of the whole sequence (Fig. S3). The similar ratio of secondary structure components was achieved through batch analysis of TAAs of Acinetobacters, implying an overall regularity in the structures of TAAs in Acinetobacters.
Structure Prediction
To determine the overall architecture of AtaA. baumannii, the protein sequence was viewed at Interpro and Pfam (Table 1). To find the location of the most conserved structural blocks, an all-against-all BLAST approach was done. All available TAAs of Moraxellacea family were blasted for clustering and for outlining the distribution of high-scoring segment pairs (HSPs) across the AtaA. baumannii sequence. The distribution of HSPs showed the conservancy of two N-terminal head domains, neck connectors, and coiled-coil regions respectively (Fig. 3). Suggesting that these domains are conserved in Moraxellaceae family. The tripartite block of Trip-ring-GIN-Neck was also found to be abundant in all TAAs; this block is repeated three times within AtaA. baumannii sequence (Fig. 4). Therefore, we attempted to build tertiary structures for aforesaid structural blocks.
Clustering analysis of TAAs of Moraxellaceae family. Each dot represents a single protein sequence. TAA sequences are grouped into six distinct collections, which are shown as clouds of assigned brown-colored circles. BLAST high-scoring segment pairs (HSPs) are shown as edges connecting dots reflecting attractive forces proportional to the negative logarithm of the HSP P value, proportional to the intensity of the gray shading for connections. The graph at the bottom left corner illustrates the distribution of HSPs through AtaA. baumannii sequence. Red asterisks show the position of head domains. Green arrows are neck connectors. The higher-level HSP scores of mentioned segments and nearby domains is evident in the graph
Since different domains of the sequence were in compliance with various homolog structures, employing multiple templates for homology modeling was a rational strategy to build a 3D model for certain parts of AtaA. baumannii. The nearly certain homolog structures to the primary sequence of AtaA. baumannii (probability greater than 95%) are presented in Fig. 4.
The most related structures were UspA1adhesin from Moraxella catarrhalis (PDB ID: 3pr7) (Agnew et al. 2011) for the head domains, AtaA C-terminal stalk from Acinetobacter sp. Tol 5 (PDB ID: 3wpa) and AtaA N-terminal half of C-terminal stalk from Acinetobacter sp. Tol 5 (PDB ID: 3wpr) (Koiwai et al. 2016) for the stalk, and Hia, a Trimeric Autotransporter from Haemophilus influenzae (PDB ID: 3emo) for the membrane anchoring region (Meng et al. 2006) (Table 3).
As illustrated in Fig. 4, two similar head domains are located after ESPR and are separated by coiled-coil regions. A trimer stalk (approximately 1050 amino acids long each monomer), is responsible for projecting globular head domains from a membrane barrel which is located at C terminus and contains 54 amino acids (each monomer). The stalk contains 4 GIN domains of ~ 50 amino acid long, three FGG motifs, two YDD domains, and a single DALL domain. Conserved residues were seen all over the structures, especially within membrane barrel and coiled-coils (Fig. 5). The head domains are globally similar to head domains of other TAAs, especially UspA (Agnew et al. 2011) which is composed of 14 repeat modules that make ladders of beta helices and all together construct a left-handed beta roll. These globular domains transit to some narrower structures (coiled-coil helices) with highly conserved neck regions which bend downward (Fig. 5) as well as the high score of B-factors (Fig S4). These features imply high levels of flexibility.
Conservancy and energetic level of protein structures. Tertiary structures are colored based on conservancy; the higher density of blue color shows a higher level of conservancy. All structures are trimers of identical chains. Energetic frustrations are presented as green (minimal frustrations) and red (maximal frustrations) lines. Focusing of maximal frustrations in certain domains are clear within the structures
Two left-handed beta roll N-terminus head domains are interleaved heads (Bassler et al. 2015) which run parallel to the fiber axis and exhibit slight structural differences. The first N-terminal head is a bit longer and more symmetric, whereas the second one shows a unique protruded peptide 610-GNKPGEG-616 at approximately half length of the second head domain (Fig. 6). The side chain of Proline in this beta-turn structure is more pronounced in the term of overhanging.
Structural alignment of two head domains. a (Algorithm: Needleman-Wunsch, Matrix: Blosum 62) structures of two head domains were aligned to show the differences between two head domains; three chains of first head domain and second head domain are illustrated by red and blue cartoons respectively. b Bottom view of the second head domain the protruded proline (P613) is evident in the image, chains are colored differently
The other globular domains (GANG, GIN, and YDD) are transversal heads, perpendicular to the fiber axis; and are class III beta hairpins (Roche et al. 2017) that are made up of tandem arrays of short repeats. GANG, GIN, and YDD are transversal heads perpendicular to the fiber axis (Fig. 5).
Energetic States of AtaA. baumannii Domains
To define the energetic states of structures, the distribution of energetic frustrations, and B-factors were determined. The highest amounts of frustrated interactions were condensed at neck connector of head domains. GIN domains and YDD domain also contain frustrated interactions (Fig. 5). Minimally frustrated interactions were distributed all over the structures (green lines in Fig. 5). The minimum and maximum frustrations were in line with B-factors (the maximum frustrations comprised higher levels of B-factors and vice versa) (Fig. S4). The high level of conservancy was also found in the highly frustrated regions.
Properties of Membrane Anchoring Region
For making a comparison, the sequence of membrane anchoring region of AtaA. baumannii was aligned with related regions of ten well-annotated TAAs from other genera. Generating a neighbor-joining tree on this alignment put AtaA. baumannii in the outgroup (Fig. 7a). Hence, the primary sequence of membrane anchoring region of TAAs in Acinetobacters seems to vary from other lineages; suggesting an interspecies modification of this gene. A comparative analysis between crystal structure of membrane channel of Hia of H. influenzae (Meng et al. 2006) and the predicted structure of AtaA.baumannii membrane barrel, was conducted to arrive at a better understanding of the properties associated with the translocator region of the protein (Fig. S5). Table 4 shows that membrane barrel of AtaA.baumannii is longer and narrower, whereas, Hia represents a wider, shorter, more symmetric, and regular cavity. The elongation of the pore of AtaA.baumannii is mainly related to the conserved Histidine residue. The lining residues through the channel in both pores are composed of polar and charged residues. In the case of AtaA. baumannii, no level of conservancy was observed through these lining residues (Fig. 7b), except for residues at the extremity of the channel.
Membrane anchoring domain properties and comparison. a A tree was built based on the alignment of AtaA. baumannii membrane anchoring region with the related region of ten well defined TAAs from other genera; the red leaf is membrane anchoring region of AtaA. baumannii which is located in an outgroup. b Visualization of the transmembrane pore in AtaA. baumannii. The translocation pore is displayed in yellow. Along the pore-axis, the positions of the pore-lining residues are identified. White colored dots (based on the conservancy, the same as Fig. 7) represent the lining residues. The lining residues of AtaA. baumannii identified as follows (with actual residue numbers): LYS1819, TYR1822, HIS1829, ARG1840, THR1842, SER1849, THR1851, SER1863, ARG1865
Discussion
Trimeric autotransporters are modular proteins with common function and highly conserved structures despite dissimilarities in primary sequences (Linke et al. 2006). For example, the sequences of membrane anchoring domains in TAAs of Acinetobacters are divergent in comparison with other TAAs. Slight differences in primary amino acid sequences may not lead to drastic differences in 3D structures and subsequently functions of the protein. This could be rooted in the fact that the functional characters are voluntarily related to the structural features of a protein rather than the primary sequence. Indeed, the protein structures are three to ten times more conserved than sequences (Illergård et al. 2009).
Homology modeling approaches are reliable methods for predicting the tertiary (Lensink et al. 2018) and the quaternary (Bertoni et al. 2017) structures of newly defined proteins. With a qualified structure in hand, many conclusions can be made on several aspects of proteins, in fact, much of our knowledge about protein functions are derived from three dimensional structures of proteins (Lensink et al. 2018).
Structural similarities were clearly observed in the built models and the previously solved structure of TAA from Acinetobacter Tol5 (Koiwai et al. 2016). Moreover, observing a similar ratio of secondary structure components in the batch analysis of Acinetobacter TAAs, suggests an overall regularity of these proteins, at least in Acinetobacter species.
Like ~ 10% of trimeric autotransporters (Dautin and Bernstein 2007), AtaA. baumannii carries an unusually extended signal peptide (Szabady et al. 2005) (ESPR, pfam13018) (Schneewind and Missiakas 2014; Szabady et al. 2005), which plays a pivotal role in the adaptation of the protein to Sec-dependent secretion machinery, which translocate the protein into the periplasm. The observed conservation for the N-terminus domain of ESPR implies an important functional role for this region. In this regard, Sijbrandi et al. proposed co-translational targeting promotion as the functional role for this domain (Sijbrandi et al. 2003).
In Enterobacteriaceae, a trimeric lipoprotein is responsible for translocating the unfolded chains of protein into the periplasm (Grin et al. 2014). The performed BLAST searches were unable to find any lipoprotein homologs in any Acinetobacter species; it seems that such lipoprotein-like proteins are absent in Acinetobacter species, suggesting that distinct proteins would assist TAAs in Acinetobacters to translocate them into the periplasm.
The primary sequence of AtaA. baumannii contains multiple repeat modules, especially in the N-terminal part. The repeat modules in the N-terminal of the sequence are related to the conserved domain of left-handed beta roll (Accession No. Cd12820) (Nummelin et al. 2004), the collagen-binding region, common in the head domain of various TAAs. A similar domain is well-defined in Yersinia enterocoloitica, YadA (Hoiczyk et al. 2000) and more closely related bacterium, Muraxella catarrhalis (UspA protein) (Agnew et al. 2011). AtaA. baumannii, harbors two structurally similar head domains. The second head (from N-terminal) comprises a short peptide that makes a beta-turn with a proline residue at the tip of the protrusion. Involvement of proline in protein–protein interaction interfaces has long been studied (Kini and Evans 1995). Especially, the existence of proline in the context of beta-turn was subjected to binding affinity analysis due to its aromatic side chain (Bhattacharyya and Chakrabarti 2003); the existence of proline residue has a positive impact on the binding affinity of the protein. Hence it can be expected that two N-terminal interleaved head domains may show different affinities or different binding properties.
The Trp-ring-GIN-NECK tripartite block that repeated three times within the architecture of AtaA. baumannii, is responsible for creating the other repetitive patterns. The structure of this tripartite block is completely similar to the head of the Bartonella Adhesin, BadA (Szczesny et al. 2008), which is suspected to be the binding site of BadA to fibronectin. Inferring through homology, it can be accomplished that AtaA. baumannii would be comprised of at least three fibronectin binding sites. Additionally, the structural components of AtaA. baumannii are similar to N-terminal head and stalk of AtaA, the TAA expressed by environmental Acinetobacter species Tol5 (Koiwai et al. 2016). Thus, the protein may also be involved in adhesion of the bacterium to abiotic surfaces (the trait that contributed to aforesaid domains of AtaA (Koiwai et al. 2016)).
One important aspect of A. baumannii lifestyle is biofilm formation (Longo et al. 2014). Biofilm formation plays an essential role in the persistence of bacteria in natural environments (Flemming et al. 2016). In the case of A. baumannii, this form of life seems to be crucial in the confrontation of the bacterium with harsh environments (such as hospital settings) (Longo et al. 2014). Along with important determinants in triggering biofilm communities by A. baumannii, such as OmpA (Gaddy et al. 2009) and biofilm-associated proteins (De Gregorio et al. 2015), TAAs are also known as surface components that are involved in biofilm formation (Bentancor et al. 2012; Ishikawa et al. 2012).
The primal requirement for TAA to involve in biofilm formation is its binding property; flexibility of highly frustrated regions of a protein has been addressed in different studies (Ferreiro et al. 2014; Parra et al. 2016). The distribution of highly frustrated contacts revealed the flexibility of related domains. The resiliency of TAAs has been previously confirmed (Geme and Cutter 2000; Koiwai et al. 2016). This property facilitates the adherence of bacterial cells to various surfaces; viewing the distribution of energetic frustrations and flexibility of regions, similar conclusions can be extrapolated from our in silico predictions. High energetic frustration is attributed to the connectors, GIN, and YDD domains. Flexible regions within the nanofiber structure of AtaA. baumannii may be the position of bending. The structural bending characteristic confers the more aggressive trait to the pathogen. Bending points could bring about the protein to bind with the involvement of multiple domains, rather than a single site. In other words, the chance of facing the binding sites to external surfaces would be increased via bending the nanofiber. It must affect the binding properties of the protein or the steps in the binding process which was also described previously for Hia, a trimeric autotransporter of Haemophilus influenzae (Meng et al. 2008). In the case of Hia, the binding is a multistep process due to the existence of multiple binding domains. GIN and C-terminal YDD heads of the AtaA. baumannii showed highly frustrated interactions, which indicate their great potential for bending and binding [the sites of protein–protein interactions have been shown to be highly frustrated (Jenik et al. 2012)]. Conversely, the distribution of minimal frustrated interactions implies a rigid stable structure for related regions.
Traditional therapeutic strategies have failed to eradicate the pathogen from infected patients or hospital settings due to the extraordinary ability of A. baumannii in adaptation and drug resistance traits. These would extremely limit the therapeutic options. Thus, vaccine development and anti-virulence drugs constitute valuable alternatives. The growing number of validated virulence protein targets with known structure, along with great power of screening algorithms, make it possible to rational design of anti-virulence drugs (Neville and Jia 2019), vaccines (Dormitzer et al. 2012; Negahdaripour et al. 2017; Nezafat et al. 2017) and diagnostics tools. Targeting the virulence factors would not affect the natural survival of pathogen, therefore, would not alter the selective evolutionary pressures that may lead to the development of drug resistance (Cegelski et al. 2008). An applicable plan in the confrontation with bacterial pathogens might be damaging the bacterial adhesion due to its preliminary role in the infection process (Cegelski et al. 2008). Development of such anti-virulence drugs requires an in-depth understanding of virulence factors (Dickey et al. 2017).
Making a complete inventory of TAAs is difficult due to the complexity of domains. Characterizing a protein from the similarity of its amino acids to the well-defined proteins is a great potential of computational methods and known as “inheritance through homology” (Lee et al. 2007). Indeed, to overcome biases, it is necessary to compare several results and linking the available data together. In the present study, we employed up to date databases and most reliable software tools; The software parameters were set to the optimized level to achieve the most accurate results. For generating tertiary structures, the best-qualified templates were selected, those that comprised a higher level of identity with the query sequence. Within generated models, the best-qualified model was selected for structural analyses. In addition, compliance of predicted secondary structures and tertiary structures was visually inspected.
Conclusion
Some structural features of AtaA. baumannii were defined by in silico tools. AtaA. baumannii could be considered as a moderate adhesin in the term of sequence length (1873 amino acids), compared to the average length of TAAs in UniProtKB database: ~ 1350. The architecture of AtaA. baumannii comprises the several well-defined structures of trimeric autotransporter family. Multiple bending and binding sites were predicted to be present in the protein structure which can assist the organism to attach to the different surfaces. Describing the sequence and structural properties of such a virulence factor would be useful in practical attempts in designing new vaccines or anti-virulence drugs.
References
Agnew C et al (2011) Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding. Proc Natl Acad Sci 108:15174–15178
Alva V, Nam S-Z, Söding J, Lupas AN (2016) The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415
Baek M, Park T, Heo L, Park C, Seok C (2017) GalaxyHomomer: a web server for protein homo-oligomer structure prediction from a monomer sequence or structure. Nucleic Acids Res 45:W320–W324
Bassler J, Alvarez BH, Hartmann MD, Lupas AN (2015) A domain dictionary of trimeric autotransporter adhesins. Int J Med Microbiol 305:265–275
Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litrán T (2012) Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960
Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T (2017) Modeling protein quaternary structure of homoand hetero-oligomers beyond binary interactions by homology. Sci Rep 7:10480
Bhattacharyya R, Chakrabarti P (2003) Stereospecific interactions of proline residues in protein structures and complexes. J Mol Biol 331:925–940
Biegert A, Söding J (2008) HHrepID: de novo protein repeat identification by probabilistic consistency. Bioinformatics 24:807–814
Bio C (2017) CLC genomics workbench. http://www.clcbio.com
Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17
Chen VB et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D 66:12–21
Consortium U (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169
Cotter SE, Surana NK, Geme JWS III (2005) Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol 13:199–205
Czibener C, Ugalde JE (2012) Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect 14:79–85
Dautin N, Bernstein HD (2007) Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol 61:89–112
De Gregorio E, Del Franco M, Martinucci M, Roscetto E, Zarrilli R, Di Nocera PP (2015) Biofilm-associated proteins: news from Acinetobacter. BMC Genomics 16:933
Diard M, Hardt W-D (2017) Evolution of bacterial virulence. FEMS Microbiol Rev 41:679–697
Dickey SW, Cheung GY, Otto M (2017) Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457
Dormitzer PR, Grandi G, Rappuoli R (2012) Structural vaccinology starts to deliver. Nat Rev Microbiol 10:807
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ferreiro DU, Komives EA, Wolynes PG (2014) Frustration in biomolecules. Q Rev Biophys 47:285–363
Finn RD et al (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199
Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563
Frickey T, Lupas A (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704
Gaddy JA, Tomaras AP, Actis LA (2009) The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160
Geme JWS, Cutter D (2000) The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J Bacteriol 182:6005–6013
Grin I et al (2014) A trimeric lipoprotein assists in trimeric autotransporter biogenesis in enterobacteria. J Biol Chem 289:7388–7398
Harding CM, Hennon SW, Feldman MF (2018) Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91
Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J (2000) Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J 19:5989–5999
Hsu L-Y, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA (2017) Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in south and southeast Asia. Clin Microbiol Rev 30:1–22
Illergård K, Ardell DH, Elofsson A (2009) Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins 77:499–508
Imperi F, Antunes L, Blom J, Villa L, Iacono M, Visca P, Carattoli A (2011) The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63:1068–1074
Ishikawa M, Nakatani H, Hori K (2012) AtaA, a new member of the trimeric autotransporter adhesins from Acinetobacter sp. Tol 5 mediating high adhesiveness to various abiotic surfaces. PLoS ONE 7:e48830
Jahangiri A, Rasooli I, Owlia P, Fooladi AAI, Salimian J (2018) An integrative in silico approach to the structure of Omp33-36 in Acinetobacter baumannii. Comput Biol Chem 72:77–86
Jenik M, Parra RG, Radusky LG, Turjanski A, Wolynes PG, Ferreiro DU (2012) Protein frustratometer: a tool to localize energetic frustration in protein molecules. Nucleic Acids Res 40:W348–W351
Kini RM, Evans HJ (1995) A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun 212:1115–1124
Koiwai K, Hartmann MD, Linke D, Lupas AN, Hori K (2016) Structural basis for toughness and flexibility in the C-terminal passenger domain of an Acinetobacter trimeric autotransporter adhesin. J Biol Chem 291:3705–3724
Lee D, Redfern O, Orengo C (2007) Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol 8:995–1005
Lensink MF, Velankar S, Baek M, Heo L, Seok C, Wodak SJ (2018) The challenge of modeling protein assemblies: the CASP12-CAPRI experiment. Proteins 86:257–273
Letunic I, Bork P (2017) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA (2006) Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol 14:264–270
Longo F, Vuotto C, Donelli G (2014) Biofilm formation in Acinetobacter baumannii. New Microbiol 37:119–127
Marchaim D et al (2017) Clinical and molecular epidemiology of Acinetobacter baumannii bloodstream infections in an endemic setting. Future Microbiol 12:271–283
Martín-Aspas A, Guerrero-Sánchez FM, García-Colchero F, Rodríguez-Roca S, Girón-González J-A (2018) Differential characteristics of Acinetobacter baumannii colonization and infection: risk factors, clinical picture, and mortality. Infect Drug Resist 11:861
Masood TB, Sandhya S, Chandra N, Natarajan V (2015) CHEXVIS: a tool for molecular channel extraction and visualization. BMC Bioinform 16:119
McConnell MJ, Actis L, Pachón J (2013) Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Meng G, Surana NK, St Geme JW, Waksman G (2006) Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J 25:2297–2304
Meng G, Geme JWS III, Waksman G (2008) Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. J Mol Biol 384:824–836
Negahdaripour M et al (2017) A novel HPV prophylactic peptide vaccine, designed by immunoinformatics and structural vaccinology approaches. Infect Genet Evol 54:402–416
Neville N, Jia Z (2019) Approaches to the structure-based design of antivirulence drugs: therapeutics for the post-antibiotic era. Molecules 24:378
Nezafat N, Eslami M, Negahdaripour M, Rahbar MR, Ghasemi Y (2017) Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol BioSyst 13:699–713. https://doi.org/10.1039/C6MB00772D
Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A (2004) The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll. EMBO J 23:701–711
Parra RG, Schafer NP, Radusky LG, Tsai M-Y, Guzovsky AB, Wolynes PG, Ferreiro DU (2016) Protein Frustratometer 2: a tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res 44:W356–W360
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Rahbar MR et al (2019) Trimeric autotransporter adhesins in Acinetobacter baumannii, coincidental evolution at work. Infect Genet Evol 71:116–127
Ritter B, Denisov AY, Philie J, Deprez C, Tung EC, Gehring K, McPherson PS (2004) Two WXXF-based motifs in NECAPs define the specificity of accessory protein binding to AP-1 and AP-2. EMBO J 23:3701–3710
Roche DB, Do Viet P, Bakulina A, Hirsh L, Tosatto SC, Kajava AV (2017) Classification of β-hairpin repeat proteins. J Struct Biol 201(2):130–138
Rodríguez-Baño J et al (2009) Long-term control of hospital-wide, endemic multidrug-resistant Acinetobacter baumannii through a comprehensive “bundle” approach. Am J Infect Control 37:715–722
Schneewind O, Missiakas D (2014) Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochem Biophys Acta 1843(8):1687–1697. https://doi.org/10.1016/j.bbamcr.2013.11.009
Sijbrandi R, Urbanus ML, Corinne M, Bernstein HD, Oudega B, Otto BR, Luirink J (2003) Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J Biol Chem 278:4654–4659
Simm D, Hatje K, Kollmar M (2015) Waggawagga: comparative visualization of coiled-coil predictions and detection of stable single α-helices (SAH domains). Bioinformatics 31:767–769
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Szabady RL, Peterson JH, Skillman KM, Bernstein HD (2005) An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA 102:221–226
Szczesny P et al (2008) Structure of the head of the Bartonella adhesin BadA. PLoS Pathog 4:e1000119
Wang S, Li W, Liu S, Xu J (2016) RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res 44:W430–W435
Webb B, Sali A (2014) Protein structure modeling with MODELLER. Curr Protoc Bioinform 47(1):5
Weidensdorfer M et al (2016) Analysis of endothelial adherence of Bartonella henselae and Acinetobacter baumannii using a dynamic human ex vivo infection model. Infect Immun 84:711–722
Wood CR, Mack LE, Actis LA (2018) An update on the Acinetobacter baumannii regulatory circuitry. Trends Microbiol 26(7):560–562
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534
Yang J, Wang Y, Zhang Y (2016) ResQ: an approach to unified estimation of B-factor and residue-specific error in protein structure prediction. J Mol Biol 428:693–701
Zimmermann L et al (2017) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430(15):2237–2243
Acknowledgements
The authors wish to thanks Pharmaceutical Science Research Center, Shiraz University of Medical Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rahbar, M.R., Zarei, M., Jahangiri, A. et al. Pierce into the Native Structure of Ata, a Trimeric Autotransporter of Acinetobacter baumannii ATCC 17978. Int J Pept Res Ther 26, 1269–1282 (2020). https://doi.org/10.1007/s10989-019-09920-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-019-09920-5