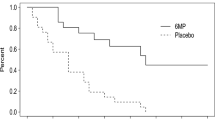
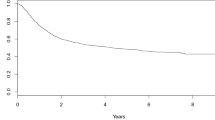
Abstract
The proportional hazards mixture cure model is a popular analysis method for survival data where a subgroup of patients are cured. When the data are interval-censored, the estimation of this model is challenging due to its complex data structure. In this article, we propose a computationally efficient semiparametric Bayesian approach, facilitated by spline approximation and Poisson data augmentation, for model estimation and inference with interval-censored data and a cure rate. The spline approximation and Poisson data augmentation greatly simplify the MCMC algorithm and enhance the convergence of the MCMC chains. The empirical properties of the proposed method are examined through extensive simulation studies and also compared with the R package “GORCure”. The use of the proposed method is illustrated through analyzing a data set from the Aerobics Center Longitudinal Study.





Similar content being viewed by others
References
Berkson J, Gage RP (1952) Survival curve for cancer patients following treatment. J Am Stat Assoc 47:501–515
Blair SN, Kampert JB, Kohl HW, Barlow CE, Macera CA, Paffenbarger RS, Gibbons LW (1996) Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. JAMA 276:205–210
Boag JW (1949) Maximum likelihood estimates of the proportion of patients cured by cancer therapy. J R Stat Soc Ser B 11:15–53
Cai B, Lin X, Wang L (2011) Bayesian proportional hazards model for current status data with monotone splines. Comput Stat Data Anal 55:2644–2651
Chen M-H, Ibrahim J, Sinha D (1999) A new Bayesian model for survival data with a surviving fraction. J Am Stat Assoc 94:909–919
Cox DR (1972) Regression models and life-tables (with discussion). J R Stat Soc Ser B 34:187–220
Dey DK, Chen M, Chang H (1997) Bayesian approach for nonlinear random effects models. Biometrics 53:1239–1252
Farewell VT (1982) The use of mixture models for the analysis of survival data with long-term survivors. Biometrics 38:1041–1046
Geisser S, Eddy WF (1979) A predictive approach to model selection. J Am Stat Assoc 74:153–160
Gelfand AE (1992) Model determination using predictive distributions with implementation via sampling-based methods (with discussion). In: Bernardo JM, Berger JO, Dawid AP, Smith AFM (eds) Bayesian statistics 4. Oxford University Press, Oxford, pp 147–167
Gilks WR, Best NG, Tan KKC (1995) Adaptive rejection Metropolis sampling within Gibbs sampling. Appl Stat 44:455–472
Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57:97–109
Ibrahim J, Chen M-H, Sinha D (2001) Bayesian semiparametric models for survival data with a cure fraction. Biometrics 57:383–388
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Kuk A, Chen C-H (1992) A mixture model combining logistic regression with proportional hazards regression. Biometrika 79:531–541
Lee DC, Sui X, Church TS, Lavie CJ, Jackson AS, Blair SN (2012) Changes in fitness and fatness on the development of cardiovascular disease risk factors: hypertension, metabolic syndrome, and hypercholesterolemia. J Am Coll Cardiol 59:665–672
Louis A (1982) Finding the observed information matrix when using the EM algorithm. J R Stat Soc Ser B Stat Methodol 44:226–233
McMahan CS, Wang L, Tebbs JM (2013) Regression analysis for current status data using the EM algorithm. Stat Med 32:4452–4466
Pan C, Cai B (2020) A Bayesian model for spatial partly interval-censored data. Commun Stat Simul Comput 51:7513–7525
Pan C, Cai B, Wang L, Lin X (2013) Bayesian semiparametric model for spatially correlated interval-censored survival data. Comput Stat Data Anal 74:198–208
Pan C, Cai B, Wang L (2015) Multiple frailty model for clustered interval-censored data with frailty selection. Stat Meth Med Res 26:1308–1322
Pan C, Cai B, Wang L (2020) A Bayesian approach for analyzing partly interval-censored data under the proportional hazards model. Stat Methods Med Res 29:3192–3204
Peng Y, Dear K (2000) A nonparametric mixture model for cure rate estimation. Biometrics 56:237–243
Peng Y, Taylor J (2011) Mixture cure model with random effects for the analysis of a multi-center tonsil cancer study. Statist Med 30:211–223
Ramsay JO (1988) Monotone regression splines in action. Stat Sci 3:425–441
Sy J, Taylor J (2000) Estimation in a Cox proportional hazards cure model. Biometrics 56:227–236
Therneau TM, Lumley T, Atkinson E, Crowson C (2021) survival: Survival analysis. https://cran.r-project.org/package=survival. R package version 3.2-13
Tsodikov A (1998) A proportional hazards model taking account of long-term survivors. Biometrics 54:1508–1516
Wang X, Wang Z (2021) EM algorithm for the additive risk mixture cure model with interval-censored data. Lifetime Data Anal 27:91–130
Wang L, McMahan CS, Hudgens MG, Qureshi ZP (2016) A flexible, computationally efficient method for fitting the proportional hazards model to interval-censored data. Biometrics 72:222–231
Xiang L, Ma X, Yau KW (2010) Mixture cure model with random effects for clustered interval-censored survival data. Stat Med 30:995–1006
Xu L, Zhang J (2010) Multiple imputation method for the semiparametric accelerated failure time mixture cure model. Comput Stat Data Anal 54:1808–1816
Xu Y, Zhao S, Hu T, Sun J (2021) Variable selection for generalized odds rate mixture cure models with interval-censored failure time data. Comput Stat Data Anal 156:107–115
Yakovlev A, Tsodikov A (1996) Stochastic models of tumor latency and their biostatistical applications. World Scientific, Singapore
Yin G, Ibrahim J (2005) A general class of Bayesian survival models with zero and nonzero cure fractions. Biometrics 61:403–412
Yin G, Ibrahim J (2005) Cure rate models: a unified approach. Can J Stat 33:559–570
Zeng D, Cai J, Shen Y (2006) Semiparametric additive risks model for interval-censored data. Stat Sin 16:287–302
Zeng D, Mao L, Lin DY (2016) Maximum likelihood estimation for semiparametric transformation models with interval-censored data. Biometrika 103:253–271
Zhang J, Peng Y (2007) A new estimation method for the semiparametric accelerated failure time mixture cure model. Statist Med 26:3157–3171
Zhou J, Zhang J, Lu W (2017) GORCure: Fit generalized odds rate mixture cure model with interval censored data. https://cran.r-project.org/package=GORCure. R package version 2.0
Zhou J, Zhang J, Lu W (2018) Computationally efficient estimation for the generalized odds rate mixture cure model with interval-censored data. J Comput Graph Stat 27:48–58
Zhou J, Zhang J, McLain AC, Cai B (2016) A multiple imputation approach for semiparametric cure model with interval censored data. Comput Stat Data Anal 99:105–114
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
After initializing values for the parameters, the proposed MCMC algorithm proceeds in the following steps.
-
1.
Let \(Z_{i} = 0\) and \(W_{i} = 0\) for all i, \(Z_{il} = 0\) and \(W_{il} = 0\) for all i and l. If \(\delta _{1i}=1\), then sample
$$\begin{aligned}{} & {} Z_{i} \sim \text {Poi}(\varLambda _0(R_{i})e^{{\varvec{\beta }}'{} \textbf{x}_{i}})1{(Z_{i}>0)}, \\{} & {} (Z_{i1},\ldots ,Z_{iK}) \sim \text {Multinomial}(Z_{i}; \gamma _{1}I_{1}(R_{i}),\ldots ,\gamma _{K}I_{K}(R_{i})). \end{aligned}$$If \(\delta _{2i}=1\), then sample
$$\begin{aligned}{} & {} W_{i} \sim \text {Poi}(\{\varLambda _0(R_{i})-\varLambda _0(L_{i})\}e^{{\varvec{\beta }}'{} \textbf{x}_{i}})1{(W_{i}>0)}, \\{} & {} (W_{i1},\ldots ,W_{iK}) \sim \text {Multinomial}(W_{i}; \gamma _{1}\{I_{1}(R_{i})-I_{1}(L_{i})\},\ldots ,\gamma _{K}\{I_{K}(R_{i})-I_{K}(L_{i})\}). \end{aligned}$$ -
2.
For \(\beta _p\) corresponding to a numeric covariate, use the ARMS algorithm to sample from its full conditional distribution
$$\begin{aligned} p(\beta _p|\cdot ) \propto [\sum _{i=1}^{n}\{x_{ip}\beta _{p}(Z_{i}\delta _{1i}+W_{i}\delta _{2i})-e^{{\varvec{\beta }}'{} \textbf{x}_{i}}(\varLambda _{0}(R_{i})(\delta _{1i}+\delta _{2i})+\varLambda _{0}(L_{i})\delta _{3i}u_{i})\}]e^{-\frac{\beta _{p}^{2}}{2\sigma _{01}^2}}. \end{aligned}$$ -
3.
For \(\beta _p\) corresponding to a dummy variable, let \(\zeta _p=\exp (\beta _p)\), sample \(\zeta _p\) from
$$\begin{aligned}{} & {} \text {Ga}(a_\zeta +\sum _{i=1}^{n}{x_{ip}(Z_{i}\delta _{1i}+W_{i}\delta _{2i})}, \\{} & {} b_\zeta +\sum _{i=1}^{n}e^{{\varvec{\beta }}_{-p}'\textbf{x}_{i,-p}}\{\varLambda _0(R_{i})(\delta _{1i}+\delta _{2i})+\varLambda _0(L_{i})\delta _{3i}u_{i}\}x_{ip}), \end{aligned}$$where \({\varvec{\beta }}_{-p}=\{\beta _{k}: k\ne p\}\) and \(\textbf{x}_{i,-p}=\{x_{ik}: k\ne p\}\).
-
4.
If \(\delta _{1i}=1\) or \(\delta _{2i}=1\) then \(u_{i}=1\) for \(i=1,\ldots ,n\). If \(\delta _{3i}=1\) then sample \(u_{i} \sim \text {Bernoulli}(p_i)\), where
$$\begin{aligned} p_{i}=E(u_{i}|\cdot )=\frac{\pi _{i}e^{-\varLambda _{u0}(L_i)\exp ({\varvec{\beta }}'{} \textbf{x}_{i})}}{(1-\pi _i)+\pi _{i}e^{-\varLambda _{u0}(L_i)\exp ({\varvec{\beta }}'{} \textbf{x}_{i})}}. \end{aligned}$$ -
5.
Sample \(\gamma _l\), \(l=1,\ldots ,K\), from
$$\begin{aligned}{} & {} \text {Ga}\left( 1+\sum _{i=1}^{n}{(Z_{il}\delta _{1i}+W_{il}\delta _{2i}}\right) , \\{} & {} \eta +\sum _{i=1}^{n}e^{{\varvec{\beta }}'{} \textbf{x}_{i}}\{I_{l}(R_{i})(\delta _{1i}+\delta _{2i})+I_{l}(L_{i})\delta _{3i}u_{i}\}). \end{aligned}$$ -
6.
Sample \(\eta\) from \(\text {Ga}(a_{\eta }+K,b_{\eta }+\sum _{l=1}^{K}\gamma _l)\).
-
7.
Sample \(\alpha _{q}\), \(q=0,\ldots ,Q\), using the ARMS algorithm from its full conditional distribution
$$\begin{aligned} p(\alpha _{q}|\cdot ) \propto \prod _{i=1}^{n}{\frac{\exp \{(\alpha _{0}+\alpha _{1}x_{i1}+\alpha _{2}x_{i2})u_{i}\}}{1+\exp (\alpha _{0}+\alpha _{1}x_{i1}+\alpha _{2}x_{i2})}}e^{-\frac{\alpha _{q}^{2}}{2\sigma _{02}^2}}. \end{aligned}$$
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, C., Cai, B. & Sui, X. A Bayesian proportional hazards mixture cure model for interval-censored data. Lifetime Data Anal 30, 327–344 (2024). https://doi.org/10.1007/s10985-023-09613-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10985-023-09613-8