Abstract
Isothermal titration calorimetry (ITC) technique was used to study the interactions of trypsin with bicyclic sunflower-derived trypsin inhibitor (SFTI-1) as well as with its new monocyclic (with disulphide bridge only) analogues (C3H5O)–SFTI-1 and (C8H15O)–SFTI-1. ITC measurements were run in 50 mM buffer solution of HEPES or Tricine of pH 8, containing 20 mM CaCl2 at 298.15 K. Based on calorimetric data, the equilibrium constants for the inhibitor–enzyme-binding processes, K, the binding stoichiometry, N (inhibitor-to-enzyme molar ratio), as well as thermodynamic parameters (ΔG, ΔH, ΔS) for the reactions were determined. The study revealed that the stoichiometry of the resulting complexes equals 1:1. The negative binding enthalpy (ΔITC H) and favourable entropy factor (TΔITC S) suggest an important contribution of hydrogen bonding as well as hydrophobic interactions to the inhibitor–enzyme affinity. Furthermore, the relationship between the modification of the peptide structure, the experimental conditions and the thermodynamic parameters has been discussed.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Serine proteases are found ubiquitously in living organisms and viral genomes [1, 2]. In humans, they play key roles in many physiological and pathological processes, such as: digestion (trypsin, chymotrypsin, pancreatic elastase), blood coagulation (thrombin, coagulation factor Xa), fibrinolysis (urokinase, plasmin), immune response (tryptase, cathepsin G, proteinase 3), type 2 diabetes (dipeptidyl peptidase IV), respiratory disease (human neutrophil elastase) and cancers [1].
The vast majority of serine proteases have chymotrypsin-like or subtilisin-like fold [3]. Their name is derived from Ser195 (chymotrypsin numbering) residue found in an enzyme active site. The Ser γ-hydroxyl group is a nucleophile that attacks a carbonyl group of the backbone of peptidic substrate and forms a tetrahedral intermediate. This action is facilitated by His57 residue acting here as a general base. The protonated His57 is stabilized by hydrogen bond formed with Asp102. All these three amino acids—Ser, His, Asp—are known as the catalytic triad or the charge relay system [1]. The acyl-enzyme intermediate is stabilized additionally by an oxyanion-binding site (also known as the oxyanion hole) formed by backbone amide groups of Gly193 and Ser195. Thereafter, the protonated His57 acting as a general acid transfers a proton to amine of the intermediate which causes expulsion of the amine fragment (the first product) as a leaving group. The covalent acyl-enzyme complex is attacked by water molecule, leading to the formation of a new tetrahedral intermediate that undergoes a subsequent breakdown. This process is also assisted by His57. Finally, a second peptide product is released, and the enzyme Ser195 is regenerated.
Serine proteases catalyse substrate hydrolysis in a highly selective, efficient and limited manner. Their proteolytic activity must be kept under tight control, and this is achieved at multiple levels: regulated expression and secretion, controlled activation of inactive pro-enzymes (zymogens), segregation within organelles (e.g. lysosomes), pH regulations as well as reversible or irreversible inhibition. Therefore, both natural and synthetic inhibitors of serine proteinase are regarded as a promising class of therapeutic agents. Some of them have been clinically approved for the treatment of various diseases [4].
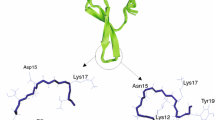
In the course of our ongoing researches on serine proteinase inhibitors, we report here the study on thermodynamic interactions of trypsin with sunflower-derived trypsin inhibitor (SFTI-1 [5]) and its new analogues (C3H5O)–SFTI-1 and (C8H15O)–SFTI-1 (Fig. 1). SFTI-1 is a backbone-cyclized peptide composed of 14 amino acids and bisected by one disulphide bridge into binding and secondary loops. SFTI-1 belongs to the well-characterized Bowman–Birk family of natural inhibitors and displays high affinity towards trypsin (based on the independent colorimetric analyses, the association constant K a = 1.1 × 1010 M−1 [6] and inhibition constant K i = 0.1 nM [5], 1 nM [7] and 13 nM [8]). Moreover, we showed that both native bicyclic SFTI-1 and its analogue deprived of the cyclic backbone, but having the disulphide bond, have comparable inhibitory activity [6]. Bowman–Birk inhibitors interact with their cognate enzymes via the standard mechanism using a common structural motif: a protease-binding loop [9, 10]. The loop is solvent-exposed, extended and complementary to the concave enzyme’s active site. The loop’s central part contains the peptide bond, which is described as a reactive site P1–P1′. According to the Schechter and Berger nomenclature [11], P1–P1′ is the scissile peptide bond and both amino acid residues P1 and P1′ interact with the corresponding enzyme’s S1 and S1′ subsides.
Chemical formulas of wild-type SFTI-1 and its two acylated analogues with disulphide bridges only. Solid lines denote covalent bonds between either amino acids or sulfhydryl groups. The P1–P1′ reactive site is located between Lys5 and Ser6 residues. (C3H5O) and (C8H15O)—propanoyl and octanoyl groups, respectively
In the current research, we analyse wild-type bicyclic SFTI-1 and its two analogues comprising the disulphide bridge only and having their N-terminal ends acylated. The acylation of a peptide N-terminal end is considered to provide enhanced stability towards proteolysis by aminopeptidase. Moreover, such modification makes peptide more hydrophobic which is known to improve cell permeability as well as may increase their therapeutic potential [12, 13].
To get better insight into the inhibitor–enzyme interaction, the isothermal titration calorimetry (ITC) technique was applied to determine the equilibrium constant for the inhibitor–enzyme-binding process, K, the binding stoichiometry, N (inhibitor-to-enzyme molar ratio), as well as thermodynamic parameters for the reactions. In particular, we aimed to investigate how the additional hydrophobic moieties introduced at the N-terminal end of the disulphide-bridged SFTI-1 would affect the inhibitor–enzyme interactions. The establishment of the enthalpy change (ΔH) and entropy change (ΔS) contributions to the formation of the inhibitor–enzyme complex may be helpful for the in-depth understanding of the relationship between the structure of the inhibitor and its physicochemical as well as biological properties.
Materials and methods
Peptide synthesis
The peptides were synthesized manually via the solid-phase approach on 2-chlorotrityl chloride resin (Calbiochem/Novabiochem) using Fmoc (fluorenyl-9-methoxycarbonyl) chemistry as was described previously [14]. The N-terminal ends of (C3H5O)–SFTI-1 and (C8H15O)–SFTI-1 were acylated with either propionic or octanoic acid (Sigma-Aldrich) in the presence of equimolar amount of N,N′-diisopropylcarbodiimide (DIPCI, GL Biochem) and 1-hydroxybenzotriazole (HOBt, GL Biochem) in dimethylformamide (DMF) for 90 min. Both peptides were cleaved form the support by a 3-h treatment with a mixture of trifluoroacetic acid (TFA)/phenol/triisopropylsilane/H2O (88:5:2:5, v/v/v/v). Under these conditions, all protecting groups were removed. In the case of wild-type SFTI-1, after completing the synthesis, the peptide was cleaved from the resin using a mixture of acetic acid/2,2,2-trifluoroethanol/methylene chloride (2:2:6, v/v/v) for 1.5 h at room temperature. Under these conditions, the cleaved peptide was protected in their side chain functions. In the next step, the head-to-tail cyclization of SFTI-1 was performed using benzotriazolyloxy-tris[pyrrolidino]-phosphonium hexafluorophosphate (PyBop)/N,N-diisopropylethylamine (molar ratio 1:2) in DMF. Next, all protecting groups were removed using TFA/phenol/triisopropylsilane/H2O (88:5:2:5, v/v/v/v). Disulphide bond formation was performed using the 0.1 M iodine solution in methanol. The peptide purity was checked by a reverse-phase high-performance liquid chromatography (RP-HPLC) on a Shimadzu Prominence UFLC equipped with a Supelco Supelcosil column (250 × 4.6 mm, 5 µm, C-8, Supelco/Sigma-Aldrich) and a UV–Vis detector. The solvent system was 0.1 % TFA (A) and 80 % acetonitrile in 0.1 % TFA (B). A linear gradient from 10 to 90 % B within 30 min, with a flow rate of 1.0 mL min−1 and monitoring set at 226 nm, was applied. The crude peptides were purified by RP-HPLC on a Beckman Gold System (Beckman) using a semi-preparative Discovery BIO Wide Pore column C-8 (250 × 10 mm, 5 μm, Supelco/Sigma-Aldrich). The solvent system was 0.1 % TFA (A) and 80 % acetonitrile in A (B). The mass spectrometry analysis of the synthesized compounds was carried out on MALDI MS (Biflex III MALDI-TOF spectrometer, Bruker Daltonics) using an α-cyano-4-hydroxycinnamic acid (CCA) and/or 2,5-dihydroxybenzoic acid (DHB) matrix.
Determination of trypsin and wild-type SFTI-1 concentrations
Bovine β-trypsin and a trypsin burst substrate nitrophenyl-4′-guanidinobenzoate (NPGB) were purchased from Sigma-Aldrich. All colorimetric measurements were performed using Cary 3E Spectrophotometer (Varian). Bovine β-trypsin solution was prepared by dissolving approximately 7 mg of the lyophilized trypsin in 1 mL of 1 mM HCl containing 20 mM CaCl2. The stock solution of bovine β-trypsin was standardized with NPGB according to Chase and Shaw method [15]. The standardized trypsin solution was used to titrate wild-type SFTI-1.
Determination of the acylated SFTI-1 analogues concentrations
The molar concentration of each peptide was determined using HPLC method as described previously [16]. Peptides were dissolved in either 50 mM Tricine (pH 8.0) or 50 mM HEPES (pH 8.0) buffers containing 20 mM CaCl2 at concentrations of about 4 mg mL−1. Equal volume of each solution was injected into the HPLC column. The peptide peak area was integrated and compared to that of the standardized wild-type SFTI-1.
Isothermal titration calorimetry (ITC)
All ITC experiments were performed at 298.15 K using an Auto-ITC isothermal titration calorimeter (MicroCal Inc., GE Healthcare, Northampton, USA) with a 1.4491 mL sample and reference cells. The reference cell contained the distilled water. All details of the measuring devices and the experimental set-up were described previously [17–19]. All reagents were dissolved directly into 50 mM buffer solution of HEPES or Tricine of pH 8.0, containing 20 mM CaCl2. The experiment consisted of injecting 10.02 μL (29 injections, 2 μL for the first injection only) of 0.23–0.27 mM buffered solution of an appropriate peptide into the reaction cell initially containing buffered solution of 0.019 mM trypsin.
A background titration, consisting of an identical titrant solution but with the buffer solution in the reaction cell only, was removed from each experimental titration to account for the heat of dilution. All the solutions were degassed prior to the titration. The titrant was injected at 5-min intervals to ensure that the titration peak returned to the baseline before the next injection. Each injection lasted 20 s. For the sake of homogeneous mixing in the cell, the stirrer speed was kept constant at 300 rpm. A calibration of the Auto-ITC calorimeter was carried out electrically by using electrically generated heat pulses. The CaCl2–EDTA titration was performed to ensure that the apparatus was working correctly, and the results (N-stoichiometry, K, ΔH) were compared with those obtained from the same samples (a test kit) at MicroCal Inc., Malvern.
Results and discussion
The stability constants (K ITC) and binding enthalpies (ΔITC H) of interactions of the inhibitors with bovine β-trypsin were obtained from ITC experiments by fitting isotherms (using nonlinear least-squares procedures) to a model that assumes a single set of identical binding sites. These thermodynamic parameters (K ITC, ΔITC H) depend on the condition under which the ITC experiments were performed (i.e. temperature, pH as well as the kind of the buffer solution). Thus, the obtained data are considered as condition-dependent parameters. Then, the conditional free energy of binding (ΔITC G) and entropy change (ΔITC S) were calculated using the standard thermodynamic relationships: ΔITC G = −RTlnK ITC = ΔITC H − TΔITC S. The results are collected in Table 1. Representative binding isotherms for interactions of the peptide tested (SFTI-1 and its acylated analogues) with trypsin are shown in Figs. 2 and 3.
The isothermal titration calorimetric data confirmed the prior knowledge [5, 20] that the stoichiometry of the resulting inhibitor–trypsin complexes equals 1:1 (SFTI-1/enzyme). The fully exposed binding loop of the bicyclic SFTI-1 possesses only one P1–P1′ reactive site with Lys residue in the P1 position that is mainly responsible for interactions with the enzyme. It was shown by Luckett et al. [5] that Lys projects into the deep S1 substrate pocket of the enzyme and makes either direct or water-mediated interactions with Ser190 and Asp189 residues found at the base of this pocket.
The data collected in Table 1 show that the thermodynamic stability of the inhibitor–enzyme complexes is comparable to each other. The change in free energy of binding (ΔITC G = −RTlnK ITC) for all the systems remains the same in the range of the experimental error. Thus, both modifications—the absence of cyclic backbone and the presence of the N-terminal hydrophobic moieties with various lengths (either propanoyl or octanoyl, Fig. 1)—do not affect the stability of the complexes studied. Furthermore, based on the ΔITC H and TΔITC S values (Table 1), it can be concluded that the formation of the complexes is both enthalpy- and entropy-driven processes. Thus, the binding of the inhibitor to enzyme is comprised of hydrogen bonding and hydrophobic interactions as indicated by the negative binding enthalpy (ΔITC H) and favourable entropy factor (TΔITC S). This finding is consistent with the data acquired from the crystallographic study [5] which showed that the interaction between native SFTI-1 and trypsin is stabilized by an extensive network of hydrogen bonds and ion pairs. On the other hand, the entropic gain may also arise from the desolvation upon the interaction of the solvent-exposed peptide loop with the concave trypsin’s active site. To assess the influence of experimental conditions on thermodynamic parameters (ΔITC H and ΔITC S), the ITC experiments were carried out in buffer solutions of the same pH, but different enthalpies of proton association with the buffer’s component (ΔBH H). This approach has been applied earlier to several protein systems including aspartic protease [21], stromelysin-1 [22] and HIV-1 protease [23] to dissect protonation contributions from binding. The enthalpy measured in the ITC experiment, ΔITC H, is the sum of all energetic effects accompanying the reaction, i.e. the enthalpy due to the heat of complex formation, Δbind H, which is independent of the kind of buffer, and the energy due to proton transfer from the reagent to the buffer [24–26]. For a given pH, the relationship between ΔITC H and the enthalpy of buffer ionization ΔBH H (BH± = B− + H+) is a straight line whose slope corresponds to the number of protons (Δn) interchanged during the reaction: ΔITC H = Δbind H + (Δn)ΔBH H. The ionization energies of the buffers used in this study are 4.9 and 7.5 kcal mol−1 for HEPES and Tricine, respectively [27]. As seen in Table 1, the measured binding enthalpies, ΔITC H, are not dependent upon the ionization enthalpy of the buffer system in which the reaction was conducted (Δn ≈ 0). Thus, it is highly probable that the inhibitor–trypsin interactions are not accompanied by a change in protonation of the reagents. For this reason, it can be supposed that the interaction studied will not be dependent upon the ionization enthalpy of the buffer used (ΔITC H ≈ Δbind H) [28].
Conclusions
The ITC technique has successfully been applied to determine the stoichiometry, binding constants (K) and thermodynamic parameters (ΔG, ΔH, ΔS) for interactions of trypsin with sunflower-derived trypsin inhibitor (SFTI-1) as well as with its new monocyclic analogues (C3H5O)–SFTI-1 and (C8H15O)–SFTI-1. It has been found that the stoichiometry of the inhibitor–enzyme complexes equals 1:1, and their stability is governed by hydrogen bonding as well as hydrophobic interactions. Carrying out the measurements in buffer solutions of equal pH but different enthalpies of ionization of its components (HEPES, Tricine) revealed that the inhibitor–trypsin interactions are not accompanied by proton transfer from the reagent to the buffer. Furthermore, it has been proved that the presence of the hydrophobic group, either propanoyl or octanoyl, at the N-terminal end of monocyclic SFTI-1 [for (C3H5O)–SFTI-1 and (C8H15O)–SFTI-1, respectively] does not affect interactions with trypsin. Thus, it can be supposed that the N-terminal end of SFTI-1 can be acylated to obtain compounds with better pharmacokinetics properties.
References
Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–24.
Page MJ, Di Cera E. Serine peptidases: classification, structure and function. Cell Mol Life Sci. 2008;65:1220–36.
Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–60.
Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68.
Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR, Brady RL. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J Mol Biol. 1999;290:525–33.
Zabłotna E, Kaźmierczak K, Jaśkiewicz A, Stawikowski M, Kupryszewski G, Rolka K. Chemical synthesis and kinetic study of the smallest naturally occurring trypsin inhibitor SFTI-1 isolated from sunflower seeds and its analogues. Biochem Biophys Res Commun. 2002;292:855–9.
Long YQ, Lee SL, Lin CY, Enyedy IJ, Wang S, Li P, Dickson RB, Roller PP. Synthesis and evaluation of the sunflower derived trypsin inhibitor as a potent inhibitor of the type II transmembrane serine protease, matriptase. Bioorg Med Chem Lett. 2001;11:2515–9.
Descours A, Moehle K, Renard A, Robinson JA. A new family of beta-hairpin mimetics based on a trypsin inhibitor from sunflower seeds. ChemBioChem. 2002;3:318–23.
Laskowski M Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626.
Krowarsch D, Cierpicki T, Jelen F, Otlewski J. Canonical protein inhibitors of serine proteases. Cell Mol Life Sci. 2003;60:2427–44.
Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–62.
Pham W, Kircher MF, Weissleder R, Tung CH. Enhancing membrane permeability by fatty acylation of oligoarginine peptides. ChemBioChem. 2004;5:1148–51.
Incani V, Lavasanifar A, Uludaǧ H. Lipid and hydrophobic modification of cationic carriers on route to superior gene vectors. Soft Matter. 2010;7:2124–38.
Dębowski D, Pikuła M, Lubos M, Langa P, Trzonkowski P, Lesner A, Łęgowska A, Rolka K. Inhibition of human and yeast 20S proteasome by analogues of trypsin inhibitor SFTI-1. PLoS One. 2014;9:e89465.
Chase T, Shaw E. Titration of trypsin, plasmin, and thrombin with p-nitrophenyl p′-guanidinobenzoate HCl. Methods Enzymol. 1970;19:20–7.
Dębowski D, Łukajtis R, Łęgowska A, Karna N, Pikuła M, Wysocka M, Maliszewska I, Sieńczyk M, Lesner A, Rolka K. Inhibitory and antimicrobial activities of OGTI and HV-BBI peptides, fragments and analogs derived from amphibian skin. Peptides. 2012;35:276–84.
Wyrzykowski D, Zarzeczańska D, Jacewicz D, Chmurzyński L. Investigation of copper(II) complexation by glycylglycine using isothermal titration calorimetry. J Therm Anal Calorim. 2011;105:1043–7.
Wyrzykowski D, Chmurzyński L. Thermodynamics of citrate complexation with Mn2+, Co2+, Ni2+ and Zn2+ ions. J Therm Anal Calorim. 2010;102:61–4.
Wyrzykowski D, Czupryniak J, Ossowski T, Chmurzyński L. Thermodynamic interactions of the alkaline earth metal ions with citric acid. J Therm Anal Calorim. 2010;102:149–54.
Korsinczky ML, Schirra HJ, Rosengren KJ, West J, Condie BA, Otvos L, Anderson MA, Craik DJ. Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J Mol Biol. 2001;311:579–91.
Xie D, Gulnik S, Collins L, Gustchina E, Suvorov L, Erickson JW. Dissection of the pH dependence of inhibitor binding energetics for an aspartic protease: direct measurement of the protonation states of the catalytic aspartic acid residues. Biochemistry. 1997;36:16166–72.
Parker MH, Lunney EA, Ortwine DF, Pavlovsky AG, Humblet C, Brouillette CG. Analysis of the binding of hydroxamic acid and carboxylic acid inhibitors to the Stromelysin-1 (matrix metalloproteinase-3) catalytic domain by isothermal titration calorimetry. Biochemistry. 1999;38:13592–601.
Velazquez-Campoy A, Kiso Y, Freire E. The binding energetics of first- and second-generation HIV-1 protease inhibitors: implications for drug design. Arch Biochem Biophys. 2001;390:169–75.
Baker BM, Murphy KP. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys J. 1996;71:2049–55.
Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–66.
Haq I, O’Brien R, Lagunavicius A, Siksnys V, Ladbury JE. Specific DNA recognition by the type II restriction endonuclease MunI: the effect of pH. Biochemistry. 2001;40:14960–7.
Goldberg RN, Kishore N, Lennen RM. Thermodynamic quantities for the ionization reactions of buffers. J Phys Chem Ref Data. 2002;31:231–70.
Gomez J, Freire E. Thermodynamic mapping of the inhibitor site of the aspartic protease endothiapepsin. J Mol Biol. 1995;252:337–50.
Acknowledgements
This work was supported by Polish National Science Center (Grant No. UMO-2011/01/D/ST5/02778).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dębowski, D., Wyrzykowski, D., Lubos, M. et al. Interactions between trypsin and its peptidic inhibitors studied by isothermal titration calorimetry (ITC). J Therm Anal Calorim 123, 807–812 (2016). https://doi.org/10.1007/s10973-015-4993-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10973-015-4993-2